7TRV
 
 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-31 | | 公開日 | 2022-02-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TWC
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DUF1479 domain-containing protein, GLYCEROL, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS
To Be Published
|
|
7TWE
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DUF1479 domain-containing protein, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid
To Be Published
|
|
8H8T
 
 | |
8H8V
 
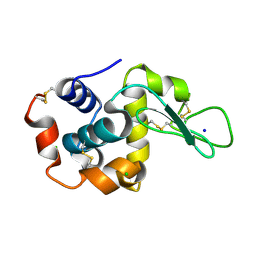 | |
8H8U
 
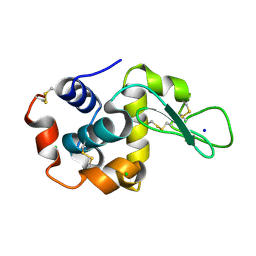 | |
8H8W
 
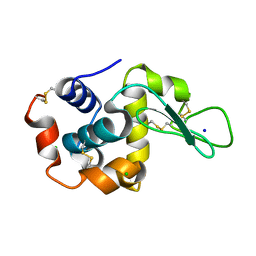 | |
1JNM
 
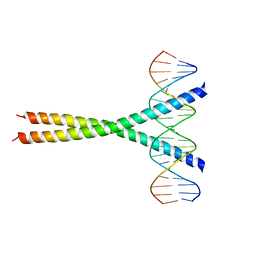 | | Crystal Structure of the Jun/CRE Complex | | 分子名称: | 5'-D(*CP*GP*TP*CP*GP*AP*TP*GP*AP*CP*GP*TP*CP*AP*TP*CP*GP*AP*CP*G)-3', PROTO-ONCOGENE C-JUN | | 著者 | Kim, Y, Podust, L.M. | | 登録日 | 2001-07-24 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Jun bZIP homodimer complexed with CRE
To be Published
|
|
1JVZ
 
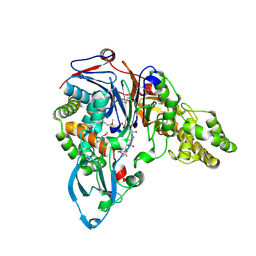 | |
1JW0
 
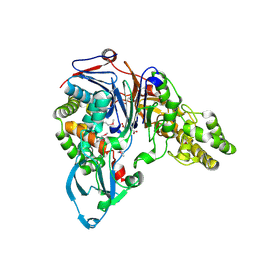 | | Structure of cephalosporin acylase in complex with glutarate | | 分子名称: | GLUTARIC ACID, cephalosporin acylase alpha chain, cephalosporin acylase beta chain | | 著者 | Kim, Y, Hol, W.G.J. | | 登録日 | 2001-09-01 | | 公開日 | 2002-09-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid and glutarate: insight into the basis of its substrate specificity
CHEM.BIOL., 8, 2001
|
|
1K77
 
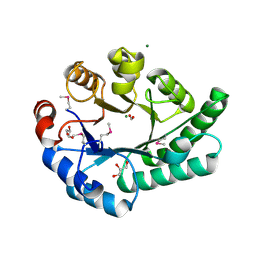 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | 分子名称: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | 著者 | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-10-18 | | 公開日 | 2002-03-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1KYT
 
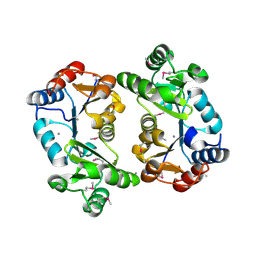 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | 分子名称: | CALCIUM ION, hypothetical protein TA0175 | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-02-05 | | 公開日 | 2003-01-21 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | 分子名称: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | 著者 | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-03-13 | | 公開日 | 2003-01-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1NJH
 
 | |
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-04 | | 公開日 | 2003-07-01 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
1NJK
 
 | | Crystal Structure of YbaW Probable Thioesterase from Escherichia coli | | 分子名称: | Hypothetical protein ybaW, IODIDE ION | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-31 | | 公開日 | 2003-07-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Hypothetical Protein YbaW
To be Published
|
|
1NE2
 
 | | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513) | | 分子名称: | FORMIC ACID, hypothetical protein ta1320 | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-10 | | 公開日 | 2003-07-01 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513)
To be Published
|
|
1NR9
 
 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | 分子名称: | MAGNESIUM ION, Protein YCGM | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-24 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-02-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
1NRI
 
 | | Crystal Structure of Putative Phosphosugar Isomerase HI0754 from Haemophilus influenzae | | 分子名称: | Hypothetical protein HI0754 | | 著者 | Kim, Y, Quartey, P, Ng, R, Zarembinski, T.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-24 | | 公開日 | 2003-07-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Hypothetical protein HI0754 from Haemophilus influenzae
To be Published
|
|
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | 分子名称: | CHLORIDE ION, SULFATE ION, yuaD protein | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-03-15 | | 公開日 | 2003-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
1OQ1
 
 | |
1P8C
 
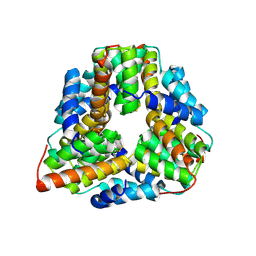 | | Crystal structure of TM1620 (APC4843) from Thermotoga maritima | | 分子名称: | conserved hypothetical protein | | 著者 | Kim, Y, Joachimiak, A, Brunzelle, J.S, Korolev, S.V, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-05-06 | | 公開日 | 2003-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure Analysis of Thermotoga maritima protein TM1620 (APC4843)
To be Published
|
|
3D0K
 
 | | Crystal structure of the LpqC, poly(3-hydroxybutyrate) depolymerase from Bordetella parapertussis | | 分子名称: | CHLORIDE ION, FORMIC ACID, Putative poly(3-hydroxybutyrate) depolymerase LpqC, ... | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-01 | | 公開日 | 2008-07-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal Structure of the LpqC, Poly(3-hydroxybutyrate) Depolymerase from Bordetella parapertussis.
To be Published
|
|
3D3O
 
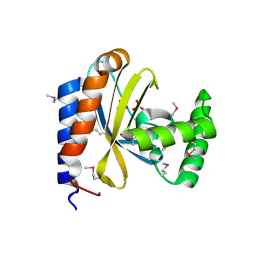 | |
3D0J
 
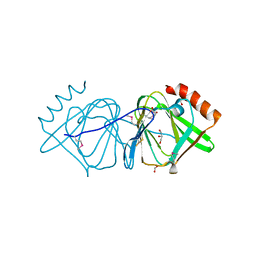 | | Crystal structure of conserved protein of unknown function CA_C3497 from Clostridium acetobutylicum ATCC 824 | | 分子名称: | FORMIC ACID, GLYCEROL, Uncharacterized protein CA_C3497 | | 著者 | Kim, Y, Bigelow, L, Clancy, S, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-01 | | 公開日 | 2008-07-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal Structure of Conserved Protein of Unknown Function CA_C3497 from Clostridium acetobutylicum ATCC 824.
To be Published
|
|
