1FQP
 
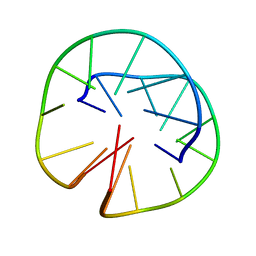 | | INTRAMOLECULAR QUADRUPLEX DNA WITH THREE GGGG REPEATS, NMR, PH 6.7, 0.1 M NA+ AND 4 MM (STRAND CONCENTRATION), 5 STRUCTURES | | Descriptor: | DNA (5'-D(GP*GP*GP*TP*TP*TP*TP*GP*GP*G)-3') | | Authors: | Keniry, M.A, Strahan, G.D, Owen, E.A, Shafer, R.H. | | Deposit date: | 1996-08-01 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Na+ form of the dimeric guanine quadruplex [d(G3T4G3)]2.
Eur.J.Biochem., 233, 1995
|
|
2AXD
 
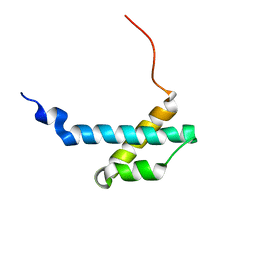 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
1BP8
 
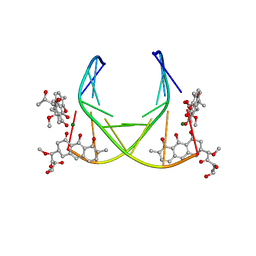 | | 4:2:1 mithramycin:Mg++:d(ACCCGGGT)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-METHYL-ANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, 5'-D(*AP*CP*CP*CP*GP*GP*GP*T)-3', ... | | Authors: | Keniry, M.A, Owen, E.A, Shafer, R.H. | | Deposit date: | 1998-08-13 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the 4:1 mithramycin:d(ACCCGGGT)2 complex: evidence for an interaction between the E saccharides
Biopolymers, 54, 2000
|
|
1DU2
 
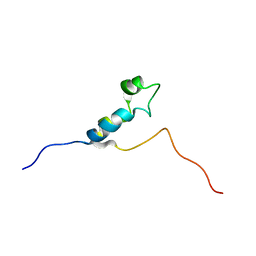 | | SOLUTION STRUCTURE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Descriptor: | DNA POLYMERASE III | | Authors: | Keniry, M.A, Berthon, H.A, Yang, J.-Y, Miles, C.S, Dixon, N.E. | | Deposit date: | 2000-01-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli.
Protein Sci., 9, 2000
|
|
3LRI
 
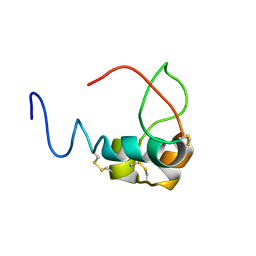 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
1JHI
 
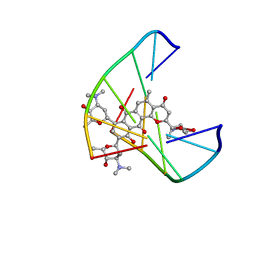 | | Solution Structure of a Hedamycin-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*(HEH)GP*GP*T)-3', HEDAMYCIN | | Authors: | Owen, E.A, Burley, G.A, Carver, J.A, Wickham, G, Keniry, M.A. | | Deposit date: | 2001-06-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the hedamycin:d(ACCGGT)2 complex by NMR and restrained molecular dynamics.
Biochem.Biophys.Res.Commun., 290, 2002
|
|
2AYA
 
 | |
2XY8
 
 | |
