6GN2
 
 | | Racemic crystal structure of A-DNA duplex formed from d(CCCGGG) in space group R3 | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), trimethylamine oxide | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2018-05-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Racemic crystal structures of A-DNA duplexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6GN3
 
 | | Racemic crystal structure of A-DNA duplex formed from d(CCCGGG) in space group P21/n | | Descriptor: | CHLORIDE ION, COBALT (II) ION, DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), ... | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2018-05-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Racemic crystal structures of A-DNA duplexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5J0E
 
 | | Crystal structures of Pribnow box consensus promoter sequence (P32) | | Descriptor: | Complementary sequence, Pribnow box consensus promoter sequence, ZINC ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5MIN
 
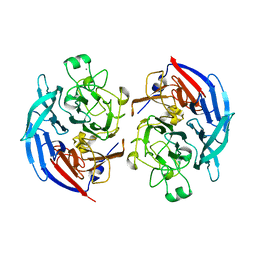 | | Apo form of the soluble PQQ-dependent Glucose Dehydrogenase from Acinetobacter calcoaceticus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Quinoprotein glucose dehydrogenase B | | Authors: | Stines-Chaumeil, C, Mavre, F, Limoges, B, Kauffmann, B, Mano, N. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Apo form of the soluble PQQ-dependent Glucose Dehydrogenase from Acinetobacter calcoaceticus
To Be Published
|
|
5HIX
 
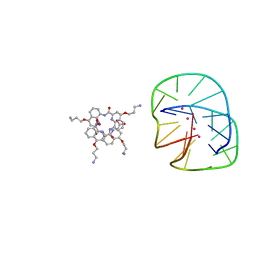 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | Descriptor: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | Authors: | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
3HCG
 
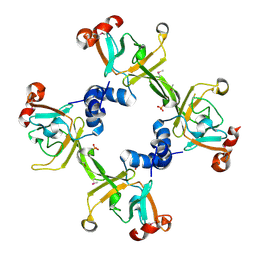 | |
8RE0
 
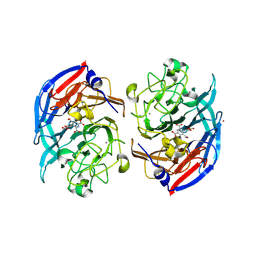 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RFK
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
2J89
 
 | | Functional and structural aspects of poplar cytosolic and plastidial type A methionine sulfoxide reductases | | Descriptor: | BETA-MERCAPTOETHANOL, METHIONINE SULFOXIDE REDUCTASE A | | Authors: | Rouhier, N, Kauffmann, B, Tete-Favier, F, Palladino, P, Gans, P, Branlant, G, Jacquot, J.P, Boschi-Muller, S. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-23 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Aspects of Poplar Cytosolic and Plastidial Type a Methionine Sulfoxide Reductases
J.Biol.Chem., 282, 2007
|
|
2FY6
 
 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
5EYQ
 
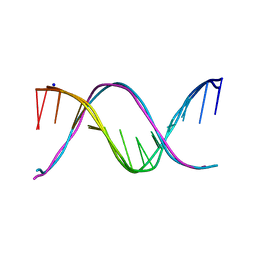 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pnna) | | Descriptor: | Complementary strand, Pribnow box template strand, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5EZF
 
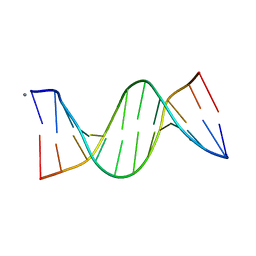 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pbca) | | Descriptor: | CALCIUM ION, Complementary strand, Pribnow box template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5F26
 
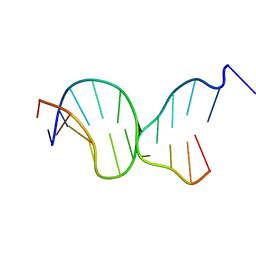 | | Crystal structures of Pribnow box consensus promoter sequence (P63) | | Descriptor: | Complementary strand, Pribnow box consensus sequence strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5EWB
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/c) | | Descriptor: | PRIBNOW BOX CONSENSUS SEQUENCE- NON-TEMPLATE STRAND, PRIBNOW BOX CONSENSUS SEQUENCE- TEMPLATE STRAND | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
3BQG
 
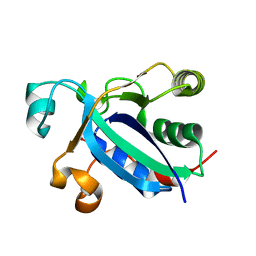 | |
3BQH
 
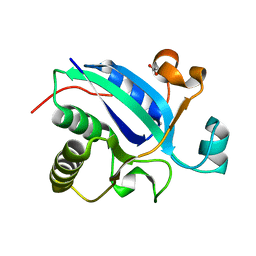 | |
3BQE
 
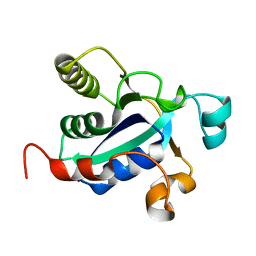 | |
3BQF
 
 | |
9GFK
 
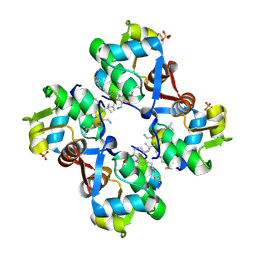 | | human MDM2 complex with stapled foldamer | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, Stapled foldamer | | Authors: | Neuville, M, Bourgeais, M, Buratto, J, Saragaglia, C, Bo, L, Mauran, L, Varajao, L, Goudreau, S.R, Kauffmann, B, Thinon, E, Pasco, M, Khatib, A.M. | | Deposit date: | 2024-08-09 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Optimal Stapling of a Helical Peptide-Foldamer Hybrid Using a C-Terminal 4-Mercaptoproline Enhances Protein Surface Recognition and Cellular Activity.
Chemistry, 31, 2025
|
|
3HCH
 
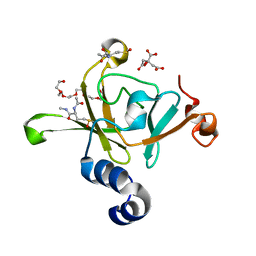 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCI
 
 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCJ
 
 | |
5ET9
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (P21/n) | | Descriptor: | BARIUM ION, Pribnow box consensus sequence- template strand, Pribnow box non-template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
