1PIH
 
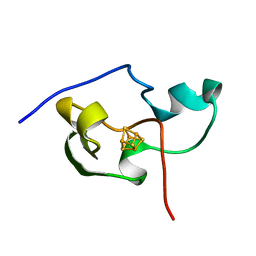 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
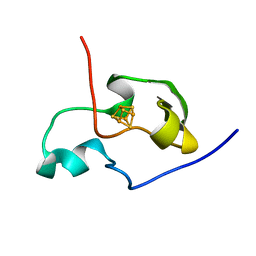 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1NEH
 
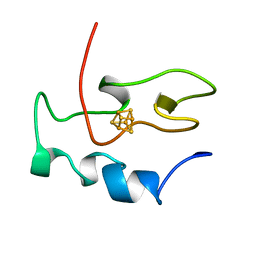 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1HRQ
 
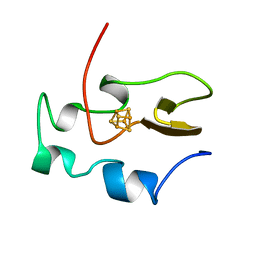 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
2BPN
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERRICYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Aguiar, A.P, Brennan, L, Xavier, A.V, Turner, D.L. | | Deposit date: | 2005-04-21 | | Release date: | 2006-03-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Tetrahaem Ferricytochrome C(3) from Desulfovibrio Vulgaris (Hildenborough) and its K45Q Mutant: The Molecular Basis of Cooperativity.
Biochim.Biophys.Acta, 1757, 2006
|
|
