1KVE
 
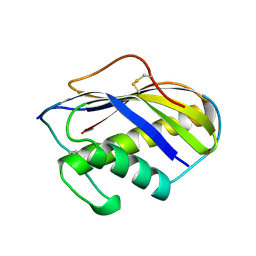 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | 分子名称: | SMK TOXIN | | 著者 | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-04-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1KVD
 
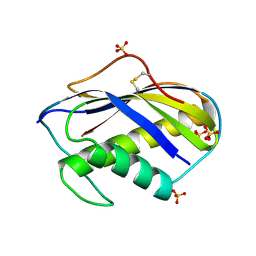 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | 分子名称: | SMK TOXIN, SULFATE ION | | 著者 | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-04-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1KVB
 
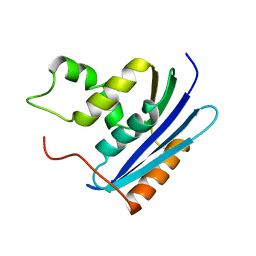 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVA
 
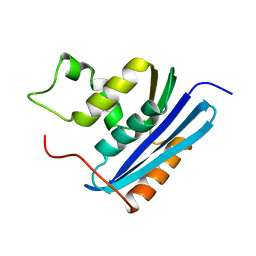 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
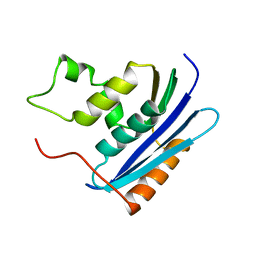 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1IU4
 
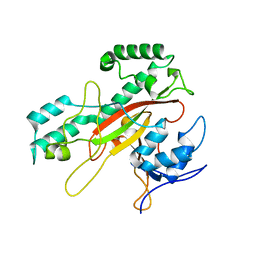 | | Crystal Structure Analysis of the Microbial Transglutaminase | | 分子名称: | microbial transglutaminase | | 著者 | Kashiwagi, T, Yokoyama, K, Ishikawa, K, Ono, K, Ejima, D, Matsui, H, Suzuki, E. | | 登録日 | 2002-02-27 | | 公開日 | 2002-08-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense
J.Biol.Chem., 277, 2002
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | 分子名称: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D48
 
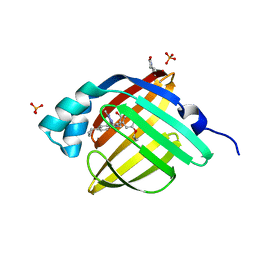 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | 分子名称: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | 分子名称: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | 分子名称: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1WP1
 
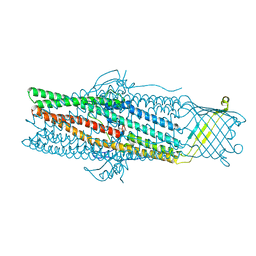 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | 分子名称: | Outer membrane protein oprM | | 著者 | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | 登録日 | 2004-08-28 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
2DPF
 
 | | Crystal Structure of curculin1 homodimer | | 分子名称: | Curculin, SULFATE ION | | 著者 | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | 登録日 | 2006-05-11 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
1VAS
 
 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | 分子名称: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | 著者 | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | 登録日 | 1995-09-08 | | 公開日 | 1996-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
5ZBC
 
 | | Crystal structure of Se-Met tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | 著者 | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | 登録日 | 2018-02-11 | | 公開日 | 2018-12-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
5ZBD
 
 | | Crystal structure of tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, TRYPTOPHAN | | 著者 | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | 登録日 | 2018-02-11 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
6ACF
 
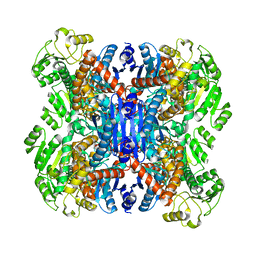 | | structure of leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | 分子名称: | Leucine dehydrogenase | | 著者 | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | 登録日 | 2018-07-26 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6ACH
 
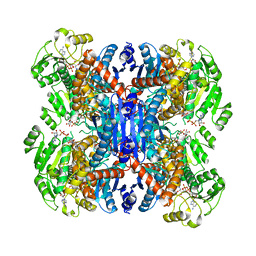 | | Structure of NAD+-bound leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | 分子名称: | Leucine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | 登録日 | 2018-07-26 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | 分子名称: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | 著者 | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | 登録日 | 2020-02-12 | | 公開日 | 2021-02-17 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
7C8I
 
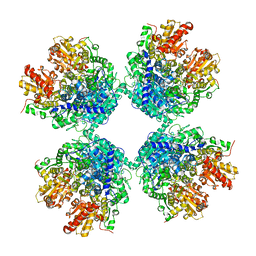 | | Ambient temperature structure of Bifidobacgterium longum phosphoketolase with thiamine diphosphate and phosphoenol pyuruvate | | 分子名称: | CALCIUM ION, PHOSPHOENOLPYRUVATE, THIAMINE DIPHOSPHATE, ... | | 著者 | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | 登録日 | 2020-06-01 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7C8H
 
 | | Ambient temperature structure of Bifidobacterium longum phosphoketolase with thiamine diphosphate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, MALONIC ACID, ... | | 著者 | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | 登録日 | 2020-06-01 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7ERU
 
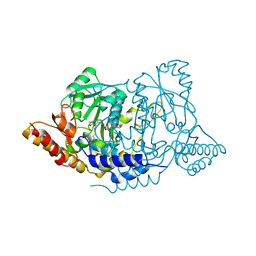 | | Crystal structure of L-histidine decarboxylase (C57S mutant) from Photobacterium phosphoreum | | 分子名称: | Histidine decarboxylase | | 著者 | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | 登録日 | 2021-05-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
7ERV
 
 | | Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum | | 分子名称: | Histidine decarboxylase, IMIDAZOLE | | 著者 | Oda, Y, Nakata, K, Yamaguchi, H, Kashiwagi, T, Miyano, H, Mizukoshi, T. | | 登録日 | 2021-05-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the enhanced thermostability of cysteine substitution mutants of L-histidine decarboxylase from Photobacterium phosphoreum.
J.Biochem., 171, 2022
|
|
3AI7
 
 | | Crystal Structure of Bifidobacterium Longum Phosphoketolase | | 分子名称: | CALCIUM ION, THIAMINE DIPHOSPHATE, Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | | 著者 | Takahashi, K, Tagami, U, Shimba, N, Kashiwagi, T, Ishikawa, K, Suzuki, E. | | 登録日 | 2010-05-10 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Bifidobacterium Longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium
Febs Lett., 584, 2010
|
|
1A99
 
 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | 分子名称: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | 著者 | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | 登録日 | 1998-04-17 | | 公開日 | 1998-10-21 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
