6D5R
 
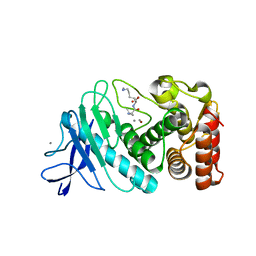 | |
6D5N
 
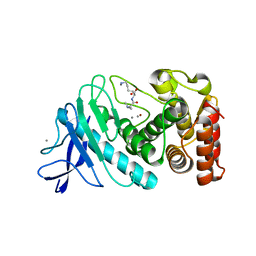 | | Hexagonal thermolysin (295) in the presence of 50% xylose | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003672 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D5U
 
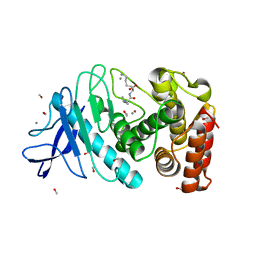 | |
6D6E
 
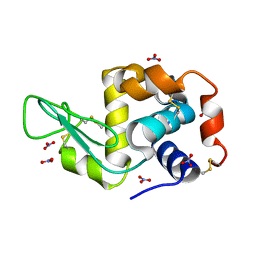 | | Triclinic lysozyme (295 K) in the presence of 47% xylose | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D5O
 
 | | Hexagonal thermolysin (295 K) in the presence of 50% DMF | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, LYSINE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00005221 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D6F
 
 | |
6D5P
 
 | |
6D6G
 
 | | Triclinic lysozyme (295 K) in the presence of 47% MPD | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D5T
 
 | | Hexagonal thermolysin cryocooled to 100 K with 50% MPD as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, LYSINE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003719 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6DZF
 
 | |
6AVL
 
 | | Orthorhombic Trypsin (295 K) in the presence of 50% xylose | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-09-02 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6P
 
 | |
6B6R
 
 | | Orthorhombic trypsin cryocooled to 100 K with 50% mpd as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-10-03 | | Release date: | 2017-11-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.00005865 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6Q
 
 | |
6B6N
 
 | | Orthorhombic trypsin (295 K) in the presence of 50% mpd | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-10-03 | | Release date: | 2017-11-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6T
 
 | |
6B6O
 
 | |
6B6S
 
 | |
3P7Q
 
 | |
3P7R
 
 | |
3P7W
 
 | |
3P7P
 
 | |
3P7V
 
 | |
3P7S
 
 | |
3P7T
 
 | |
