8KE8
 
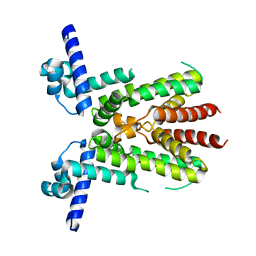 | | Crystal structure of TetR-type transcriptional factor NalC from P. aeruginosa | | Descriptor: | NalC | | Authors: | Lee, J.Y, Jeong, K.H, Ko, J.H, Son, S.B. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the transcriptional regulator NalC, a key component of the MexAB-OprM efflux pump system, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 679, 2023
|
|
7WB3
 
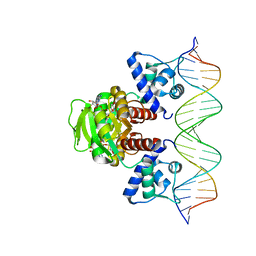 | | Crystal structure of T. maritima Rex in ternary complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*GP*AP*GP*AP*AP*AP*TP*TP*TP*AP*TP*CP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*TP*GP*AP*TP*AP*AP*AP*TP*TP*TP*CP*TP*CP*AP*AP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lee, J.Y, Jeong, K.H, Lee, H.J, Park, Y.W. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA.
Int J Mol Sci, 23, 2022
|
|
7CV2
 
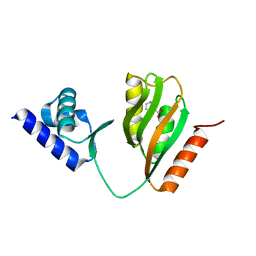 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | Descriptor: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
7CV0
 
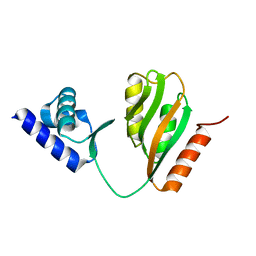 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
6KTB
 
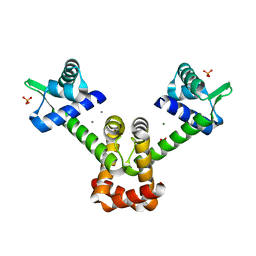 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | HTH-type transcriptional regulator MntR, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
6KTA
 
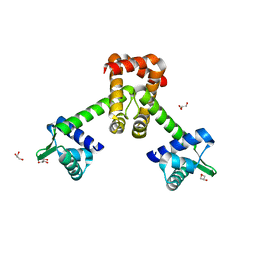 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MntR | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
7D1I
 
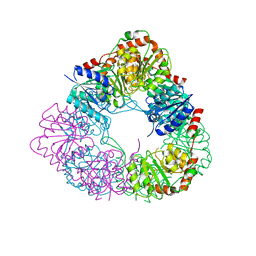 | | Crystal structure of acinetobacter baumannii MurG | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | | Authors: | Park, H.H, Jeong, k.H. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
Iucrj, 8, 2021
|
|
7D27
 
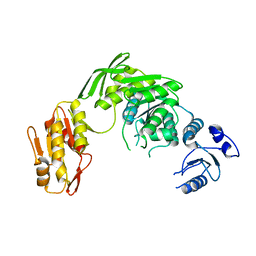 | |
