1BJO
 
 | |
1BJN
 
 | |
1MAP
 
 | |
1MAQ
 
 | |
1NPC
 
 | |
1OAT
 
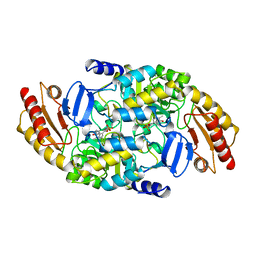 | | ORNITHINE AMINOTRANSFERASE | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shen, B.W, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1997-03-26 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human recombinant ornithine aminotransferase.
J.Mol.Biol., 277, 1998
|
|
1OXO
 
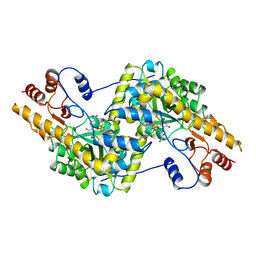 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, OPEN CONFORMATION | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Hohenester, E, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1995-12-23 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1OXP
 
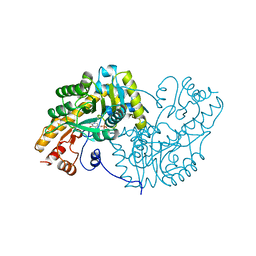 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, CLOSED CONFORMATION | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Hohenester, E, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1995-12-23 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1DGD
 
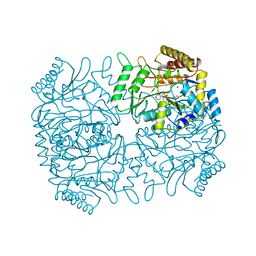 | |
1DGE
 
 | |
1F9M
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (SHORT FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-11 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB0
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (REDUCED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB6
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (OXIDIZED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FAA
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (LONG FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-13 | | Release date: | 2000-09-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1I4N
 
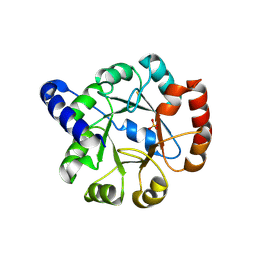 | | CRYSTAL STRUCTURE OF INDOLEGLYCEROL PHOSPHATE SYNTHASE FROM THERMOTOGA MARITIMA | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Knoechel, T, Pappenberger, A, Jansonius, J.N, Kirschner, K. | | Deposit date: | 2001-02-22 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of indoleglycerol-phosphate synthase from Thermotoga maritima. Kinetic stabilization by salt bridges.
J.Biol.Chem., 277, 2002
|
|
1IGS
 
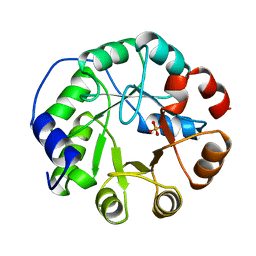 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1995-08-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 A structure of indole-3-glycerol phosphate synthase from the hyperthermophile Sulfolobus solfataricus: possible determinants of protein stability.
Structure, 3, 1995
|
|
1AMA
 
 | |
1A53
 
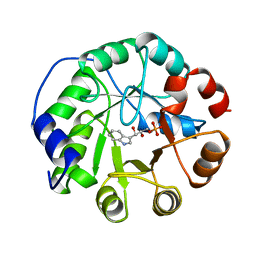 | | COMPLEX OF INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS WITH INDOLE-3-GLYCEROLPHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1998-02-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
1BT4
 
 | |
1D7S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH DCS | | Descriptor: | D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D6S
 
 | | CRYSTAL STRUCTURE OF THE K41A MUTANT OF O-ACETYLSERINE SULFHYDRYLASE COMPLEXED IN EXTERNAL ALDIMINE LINKAGE WITH METHIONINE | | Descriptor: | METHIONINE, O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Tai, C.H, Ristroph, C.M, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-10-15 | | Release date: | 2000-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 291, 1999
|
|
1D7V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1DKA
 
 | | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | | Descriptor: | 2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, ... | | Authors: | Toney, M.D, Hohenester, E, Jansonius, J.N. | | Deposit date: | 1993-06-18 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites.
Science, 261, 1993
|
|
