1GMH
 
 | |
1GQR
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH RIVASTIGMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE, ... | | Authors: | Raves, M.L, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1HCZ
 
 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
1GQS
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH NAP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE | | Authors: | Bar-on, P, Millard, C.B, Harel, M, Dvir, H, Enz, A, Sussman, J.L, Silman, I. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1I7B
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND S-ADENOSYLMETHIONINE METHYL ESTER | | Descriptor: | 1,4-DIAMINOBUTANE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, S-ADENOSYLMETHIONINE DECARBOXYLASE BETA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I72
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-07 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7C
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH METHYLGLYOXAL BIS-(GUANYLHYDRAZONE) | | Descriptor: | 1,4-DIAMINOBUTANE, METHYLGLYOXAL BIS-(GUANYLHYDRAZONE), S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7M
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE | | Descriptor: | 1,4-DIAMINOBUTANE, 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I79
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I9K
 
 | |
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
3TP6
 
 | |
3HXQ
 
 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | Descriptor: | Aptamer ARC1172, von Willebrand Factor | | Authors: | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | Deposit date: | 2009-06-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
3TPJ
 
 | | APO structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U1T
 
 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TP5
 
 | |
3TPL
 
 | | APO Structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TU3
 
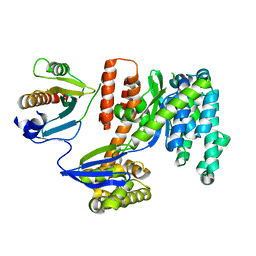 | | 1.92 Angstrom resolution crystal structure of the full-length SpcU in complex with full-length ExoU from the type III secretion system of Pseudomonas aeruginosa | | Descriptor: | ExoU, ExoU chaperone | | Authors: | Halavaty, A.S, Borek, D, Otwinowski, Z, Minasov, G, Veesenmeyer, J.L, Tyson, G, Shuvalova, L, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Type III Secretion Effector Protein ExoU in Complex with Its Chaperone SpcU.
Plos One, 7, 2012
|
|
3TZ4
 
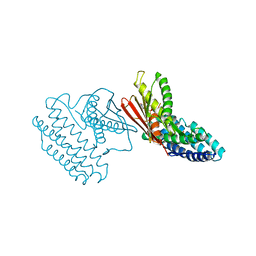 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex with ADP | | Descriptor: | (2S)-2-chloro-4-methylpentanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TP8
 
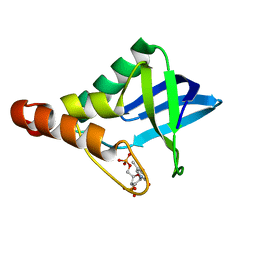 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36E at cryogenic temperature
To be Published
|
|
3TWE
 
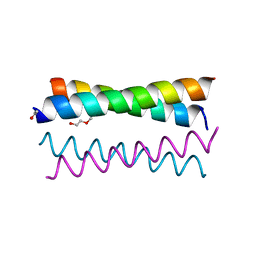 | | Crystal Structure of the de novo designed peptide alpha4H | | Descriptor: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWG
 
 | |
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | Descriptor: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UFM
 
 | |
