1OS8
 
 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1OSS
 
 | | T190P STREPTOMYCES GRISEUS TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-20 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
4QDD
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with 1,4-30Q-CoA | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDC
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with FE2/S2 (INORGANIC) CLUSTER | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QCK
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis in complex with 4-androstene-3,17-dione | | Descriptor: | 3-ketosteroid-9-alpha-monooxygenase oxygenase subunit, 4-ANDROSTENE-3-17-DIONE, FE (III) ION, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4QDF
 
 | | Crystal structure of apo KshA5 and KshA1 in complex with 1,4-30Q-CoA from R. rhodochrous | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
4DID
 
 | |
1JTD
 
 | | Crystal structure of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase | | Descriptor: | CALCIUM ION, TEM-1 beta-lactamase, beta-lactamase inhibitor protein II | | Authors: | Lim, D.C, Park, H.U, De Castro, L, Kang, S.G, Lee, H.S, Jensen, S, Lee, K.J, Strynadka, N.C.J. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO6
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CB2
 
 | | Crystal structure of Escherichia coli UppP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | Authors: | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
6CO9
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6COJ
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2WQP
 
 | | Crystal structure of sialic acid synthase NeuB-inhibitor complex | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, 5-(ACETYLAMINO)-3,5-DIDEOXY-2-O-PHOSPHONO-D-ERYTHRO-L-MANNO-NONONIC ACID, ... | | Authors: | Liu, F, Lee, H.J, Strynadka, N.C.J, Tanner, M.E. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Inhibition of Neisseria Meningitidis Sialic Acid Synthase by a Tetrahedral Intermediate Analog.
Biochemistry, 48, 2009
|
|
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
4X7P
 
 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XP8
 
 | |
4X6L
 
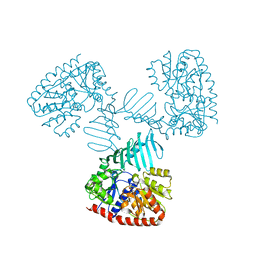 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3UJZ
 
 | |
1JTG
 
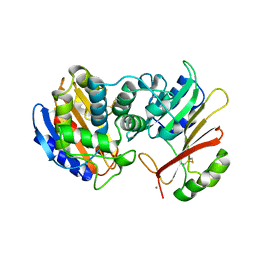 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
4G1I
 
 | |
3J1W
 
 | |
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
1JHE
 
 | | LEXA L89P Q92W E152A K156A MUTANT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, J Strynadka, N.C. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
3GMW
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | Descriptor: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
