6KIL
 
 | | N21Q mutant thioredoxin from Halobacterium salinarum NRC-1 | | Descriptor: | Thioredoxin | | Authors: | Arai, S, Shibazaki, C, Shimizu, R, Adachi, M, Ishibashi, M, Tokunaga, H, Tokunaga, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic mechanism and evolutional characteristics of thioredoxin from Halobacterium salinarum NRC-1.
Acta Crystallogr.,Sect.D, 76, 2020
|
|
2AZ1
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum | | Descriptor: | CALCIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
2AZ3
 
 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum in complex with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS5
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VGS
 
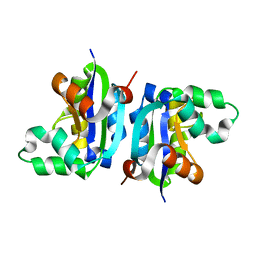 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGT
 
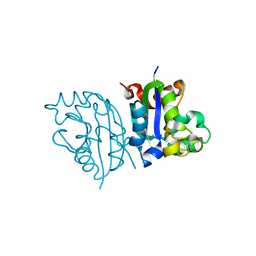 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGV
 
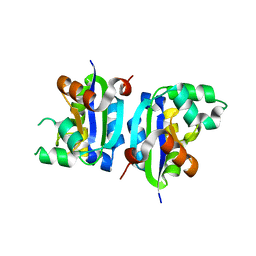 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3WBH
 
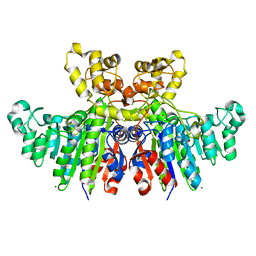 | | Structural characteristics of alkaline phosphatase from a moderately halophilic bacteria Halomonas sp.593 | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Ishibashi, M, Matsumoto, F, Tamada, T, Tokunaga, H, Tokunaga, M, Kuroki, R. | | Deposit date: | 2013-05-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characteristics of alkaline phosphatase from the moderately halophilic bacterium Halomonas sp. 593.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3VGU
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
2D9Q
 
 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
