5HT9
 
 | |
7B6E
 
 | | Drosophila melanogaster TRAPP C8 subunits region 355 to 661 | | Descriptor: | FI18195p1 | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6R
 
 | | Drosophila melanogaster TRAPPIII partial complex: core plus C8 and C11 attached region | | Descriptor: | FI18195p1, GEO08327p1, Probable trafficking protein particle complex subunit 2, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6H
 
 | | Drosophila melanogaster TRAPP C11 subunits region 1 to 718 | | Descriptor: | Trafficking protein particle complex subunit 11 | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6D
 
 | | Drosophila melanogaster TRAPPCore (C1, C2, C2L, C3a/b, C4, C5, C6 subunits) | | Descriptor: | GEO08327p1, Probable trafficking protein particle complex subunit 2, TRAPPC2L, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B70
 
 | | TRAPPCore plus C8 (355-596) and C11 (1-718) from MiniTRAPPIII | | Descriptor: | FI18195p1, GEO08327p1, Probable trafficking protein particle complex subunit 2, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6X
 
 | |
7B6Y
 
 | |
7B6Z
 
 | |
2GS4
 
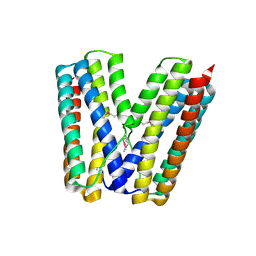 | | The crystal structure of the E.coli stress protein YciF. | | Descriptor: | Protein yciF | | Authors: | Hindupur, A, Liu, D, Zhao, Y, Bellamy, H.D, White, M.A, Fox, R.O. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the E. coli stress protein YciF.
Protein Sci., 15, 2006
|
|
5EE5
 
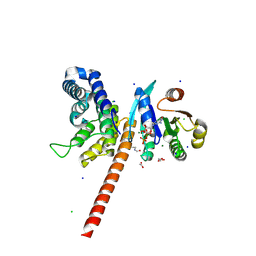 | | Structure of human ARL1 in complex with the DCB domain of BIG1 | | Descriptor: | ACETATE ION, ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, ... | | Authors: | Galindo, A, Soler, N, Munro, S. | | Deposit date: | 2015-10-22 | | Release date: | 2016-07-06 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi.
Cell Rep, 16, 2016
|
|
1MXD
 
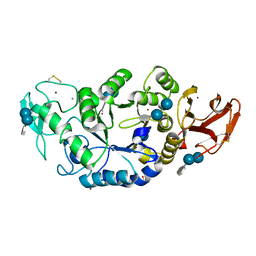 | | Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MXG
 
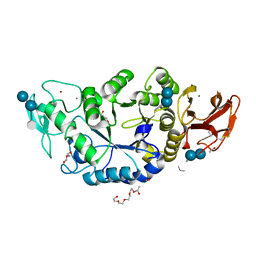 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MWO
 
 | | Crystal Structure Analysis of the Hyperthermostable Pyrocoocus woesei alpha-amylase | | Descriptor: | CALCIUM ION, ZINC ION, alpha amylase | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-09-30 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
5FGO
 
 | |
3ZS0
 
 | | Human Myeloperoxidase inactivated by TX2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-FLUOROBENZYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
3ZS1
 
 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
4UAS
 
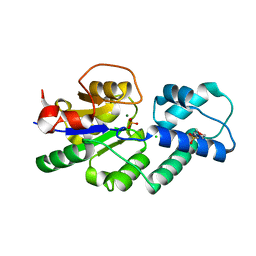 | | Crystal structure of CbbY from Rhodobacter sphaeroides in complex with phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAT
 
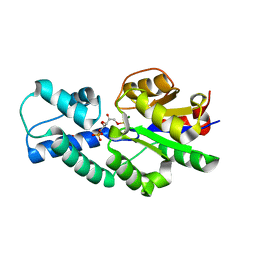 | | Crystal structure of CbbY (mutant D10N) from Rhodobacter sphaeroides in complex with Xylulose-(1,5)bisphosphate, crystal form I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Protein CbbY, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAV
 
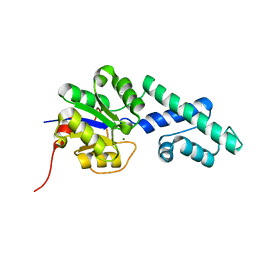 | | Crystal structure of CbbY (AT3G48420) from Arabidobsis thaliana | | Descriptor: | Haloacid dehalogenase-like hydrolase domain-containing protein At3g48420, MAGNESIUM ION | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAU
 
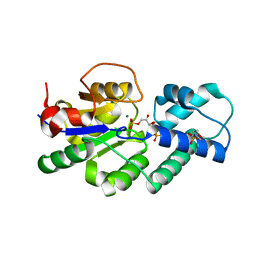 | | Crystal structure of CbbY (mutant D10N) from Rhodobacter sphaeroides in complex with Xylulose-(1,5)bisphosphate, crystal form II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Protein CbbY, ... | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAR
 
 | | Crystal structure of apo-CbbY from Rhodobacter sphaeroides | | Descriptor: | GLYCEROL, Protein CbbY | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
2VH1
 
 | | Crystal structure of bacterial cell division protein FtsQ from E.coli | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Naninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
2VH2
 
 | | Crystal structure of cell divison protein FtsQ from Yersinia enterecolitica | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Nanninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
7OF2
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with GTPBP6. | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
