6T8H
 
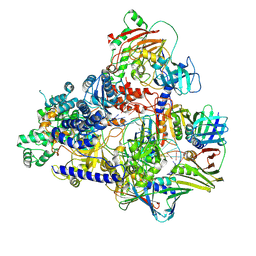 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7Y
 
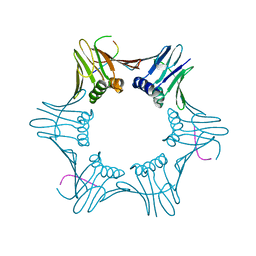 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7X
 
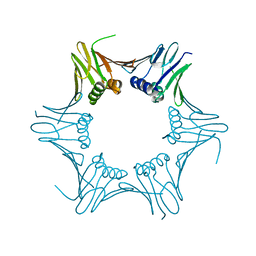 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
