8JRM
 
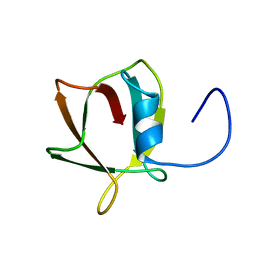 | |
8JRI
 
 | |
8JRT
 
 | |
8JTI
 
 | |
8K0G
 
 | |
1SIY
 
 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
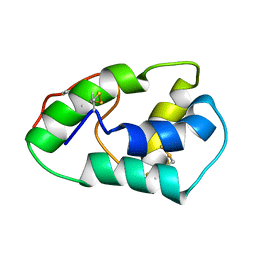
7CEM
 
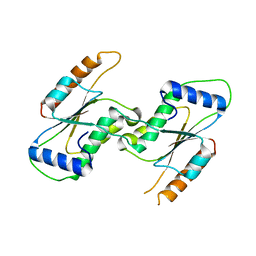 | | Crystal Structure of YbeA CP74 W7F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Wu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4251 Å) | | Cite: | Crystal Structure of YbeA CP74 W7F
To Be Published
|
|
7CFY
 
 | | Crystal Structure of YbeA CP74 W48F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4051 Å) | | Cite: | Crystal Structure of YbeA CP74 W48F
To Be Published
|
|
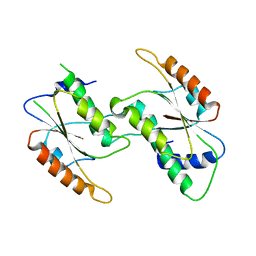
7CF7
 
 | | Crystal Structure of YbeA CP74 W72F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6178 Å) | | Cite: | Crystal Structure of YbeA CP74 W72F
To Be Published
|
|
7Y39
 
 | | Ubiquitin-like domain of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insight into ZFAND1 and p97 Interaction
To Be Published
|
|
7EJ5
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb45, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45
To Be Published
|
|
7EJ4
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb-25, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25
To be published
|
|
7D51
 
 | |
7V8A
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
To Be Published
|
|
7V8C
 
 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation
To Be Published
|
|
7V78
 
 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1
To Be Published
|
|
7V7A
 
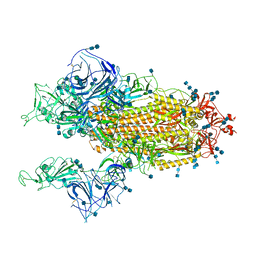 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation
To Be Published
|
|
7V7U
 
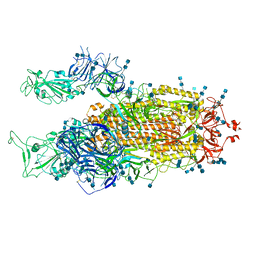 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2
To Be Published
|
|
7V7E
 
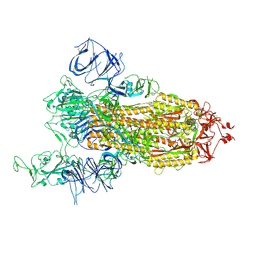 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1
To Be Published
|
|
7V82
 
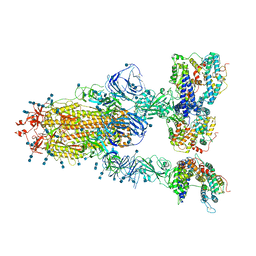 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1
To Be Published
|
|
7V7H
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1
To Be Published
|
|
7V7R
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4
To Be Published
|
|
7V7D
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), all RBD-down conformation
To Be Published
|
|
7V7S
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 5
To Be Published
|
|
7V79
 
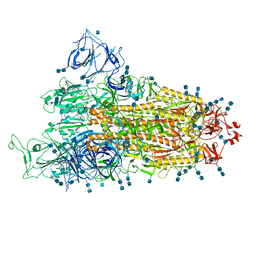 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 2
To Be Published
|
|
