6P0F
 
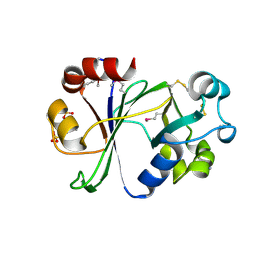 | | N-terminal domain of Thermococcus Gammatolerans McrB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, GTPase subunit of restriction endonuclease, ... | | Authors: | Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | The structure of theThermococcus gammatoleransMcrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.
J.Biol.Chem., 295, 2020
|
|
6P0G
 
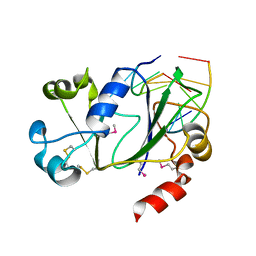 | | N-terminal domain of Thermococcus Gammatolerans McrB bound to m5C DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*AP*CP*CP*GP*G)-3'), GTPase subunit of restriction endonuclease | | Authors: | Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of theThermococcus gammatoleransMcrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.
J.Biol.Chem., 295, 2020
|
|
6N0S
 
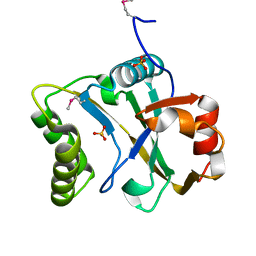 | | N-terminal domain of Staphylothermus marinus McrB | | Descriptor: | ATPase associated with various cellular activities, AAA_5, SULFATE ION | | Authors: | Hosford, C.J, Niu, Y, Chappie, J.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The N-terminal domain of Staphylothermus marinus McrB shares structural homology with PUA-like RNA binding proteins.
J.Struct.Biol., 211, 2020
|
|
6C5D
 
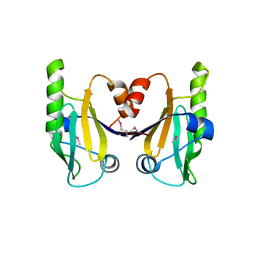 | |
6NJX
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with mercury | | Descriptor: | IODIDE ION, MERCURY (II) ION, Xcc_ctr_Hg | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NK8
 
 | | C-terminal region of the Burkholderia pseudomallei OLD protein | | Descriptor: | Class 2 OLD family nuclease, MAGNESIUM ION | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NJW
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with platinum | | Descriptor: | IODIDE ION, MAGNESIUM ION, PLATINUM (II) ION, ... | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NJV
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with iodine | | Descriptor: | IODIDE ION, MAGNESIUM ION, Xcc_CTR_I | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6UT6
 
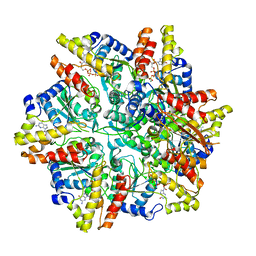 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT3
 
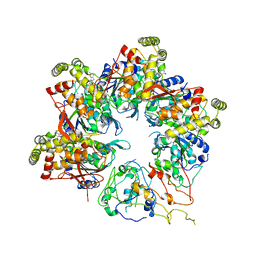 | |
6UT4
 
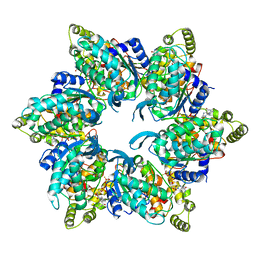 | | Cryo-EM structure of the asymmetric AAA+ domain hexamer from Thermococcus gammatolerans McrB | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, MAGNESIUM ION | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT5
 
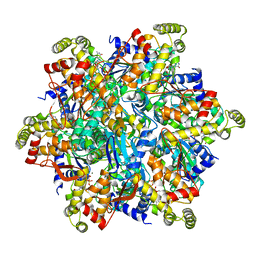 | | Cryo-EM structure of the Thermococcus gammatolerans McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT7
 
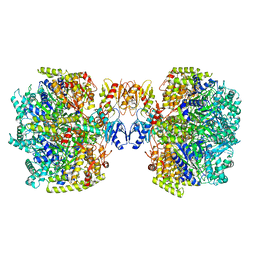 | | Fitted model for the tetradecameric assembly of Thermococcus gammatolerans McrB AAA+ hexamers with bound McrC | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT8
 
 | | Refined half-complex from tetradecameric assembly of Thermococcus gammatolerans McrB AAA+ hexamers with bound McrC | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6DJQ
 
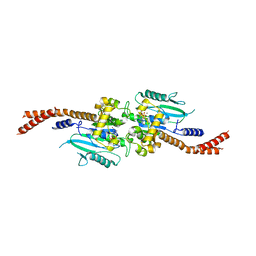 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DEF
 
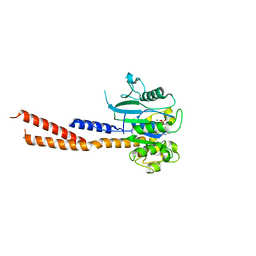 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
8EM1
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA Unbound | | Descriptor: | 1,2-ETHANEDIOL, PaqCI, DNA Unbound | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
8EPX
 
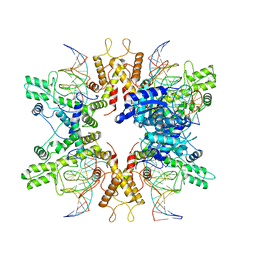 | | Type IIS Restriction Endonuclease PaqCI, DNA bound | | Descriptor: | CALCIUM ION, DNA 1a, DNA 1b, ... | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
7ULN
 
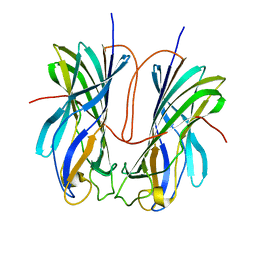 | | Turnip yellows virus N-terminal readthrough domain | | Descriptor: | Minor capsid protein P3-RTD | | Authors: | Schiltz, C.J, Chappie, J.S. | | Deposit date: | 2022-04-05 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Polerovirus N-terminal readthrough domain structures reveal molecular strategies for mitigating virus transmission by aphids
Nat Commun, 13, 2022
|
|
7ULO
 
 | |
8TWQ
 
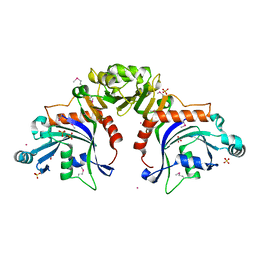 | | Structure of bacteriophage lambda RexA protein | | Descriptor: | CADMIUM ION, Protein rexA, SULFATE ION | | Authors: | Adams, M.C, Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of bacteriophage lambda RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins.
Nucleic Acids Res., 52, 2024
|
|
6VJF
 
 | | The P-Loop K to A mutation of C. therm Vps1 GTPase-BSE | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Putative sorting protein Vps1 | | Authors: | Tornabene, B.A, Varlakhanova, N.V, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural and functional characterization of the dominant negative P-loop lysine mutation in the dynamin superfamily protein Vps1.
Protein Sci., 29, 2020
|
|
6DI7
 
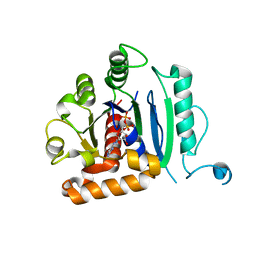 | | Vps1 GTPase-BSE fusion complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative sorting protein | | Authors: | Varlakhanova, N.V, Brady, T.M, Ford, M.G.J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
