5Z09
 
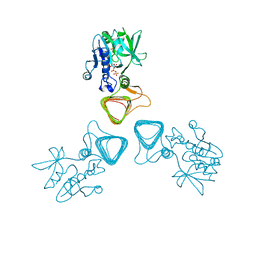 | | ST0452(Y97N)-UTP binding form | | 分子名称: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | 著者 | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | 登録日 | 2017-12-19 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
5Z0A
 
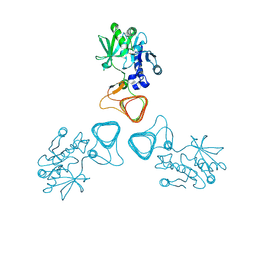 | | ST0452(Y97N)-GlcNAc binding form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | 著者 | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | 登録日 | 2017-12-19 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
5DGQ
 
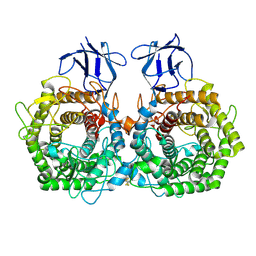 | |
5DGR
 
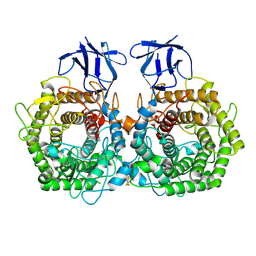 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | 著者 | Suzuki, K, Honda, Y, Fushinobu, S. | | 登録日 | 2015-08-28 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
2DRR
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRS
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263S mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRO
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263C mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRQ
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263G mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2006-06-12 | | 公開日 | 2006-06-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
1WU5
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase complexed with xylose | | 分子名称: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose, ... | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU4
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | 分子名称: | GLYCEROL, NICKEL (II) ION, xylanase Y | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU6
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase E70A mutant complexed with xylobiose | | 分子名称: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | 著者 | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | 登録日 | 2004-12-01 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | 分子名称: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | 著者 | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | 登録日 | 2009-06-22 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
1V7V
 
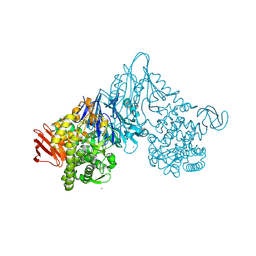 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | 分子名称: | CALCIUM ION, chitobiose phosphorylase | | 著者 | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | 登録日 | 2003-12-24 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7W
 
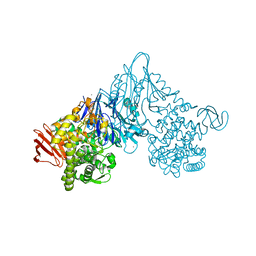 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | 登録日 | 2003-12-24 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7X
 
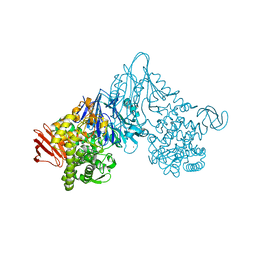 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | 登録日 | 2003-12-24 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
3UES
 
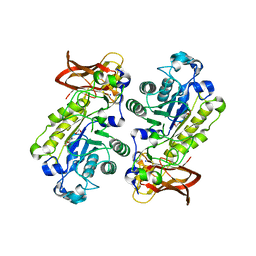 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | 分子名称: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | 著者 | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | 登録日 | 2011-10-31 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | 著者 | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | 登録日 | 2011-10-31 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
5GQC
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQG
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQF
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
