9D37
 
 | | Nonactive state of Gly-,Glu- bound GluN1a-2B-2D NMDAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
9D38
 
 | | Open state of Gly-,Glu-,EU1622-240 bound GluN1a-2B-2D NMDAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
9D3B
 
 | | Gly-,Glu-,(S)-DQP-997-74 bound GluN1a-2B-2D NMDAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{(3R,5S)-5-(4-chlorophenyl)-3-[4-(4-chlorophenyl)-2-oxo-1,2-dihydroquinolin-3-yl]pyrazolidin-1-yl}-3,3-difluoro-4-oxobutanoic acid, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
9D39
 
 | | Gly-,PPDA- bound GluN1a-2B-2D NMDAR | | Descriptor: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
9D3A
 
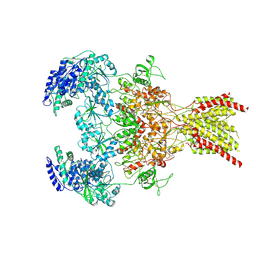 | | Nonactive state of Gly-,Glu- bound GluN1a-2B-2D NMDAR (Low-res) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
9D3C
 
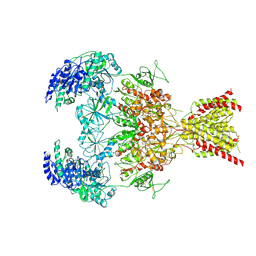 | | Gly-,Glu-,(S)-(+)-ketamine bound GluN1a-2B-2D NMDAR | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hyunook, K, Hiro, F. | | Deposit date: | 2024-08-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for channel gating and blockade in tri-heteromeric GluN1-2B-2D NMDA receptor.
Neuron, 113, 2025
|
|
1IVZ
 
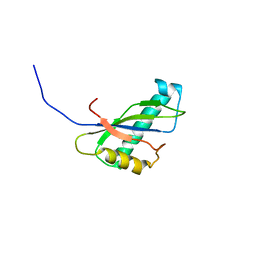 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
8BXJ
 
 | |
8C2Q
 
 | | Silver ion-bound structure of the silver specific chaperone SilF needed for bacterial silver resistance | | Descriptor: | Copper ABC transporter substrate-binding protein, SILVER ION | | Authors: | Monneau, Y.R, Walker, O, Hologne, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | The battle for silver binding: How the interplay between the SilE, SilF, and SilB proteins contributes to the silver efflux pump mechanism.
J.Biol.Chem., 299, 2023
|
|
