3BPP
 
 | |
3BK6
 
 | |
2DEO
 
 | |
3VIV
 
 | | 1510-N membrane-bound stomatin-specific protease K138A mutant in complex with a substrate peptide | | Descriptor: | 441aa long hypothetical nfeD protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yokoyama, H, Matsui, I, Fujii, S. | | Deposit date: | 2011-10-12 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a membrane stomatin-specific protease in complex with a substrate Peptide
Biochemistry, 51, 2012
|
|
1ZNT
 
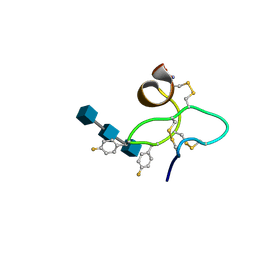 | | 18 NMR structures of AcAMP2-Like Peptide with non Natural Fluoroaromatic Residue (AcAMP2F18Pff/Y20Pff) complex with N,N,N-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Aboitiz, N, Freire, F, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-12-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZUV
 
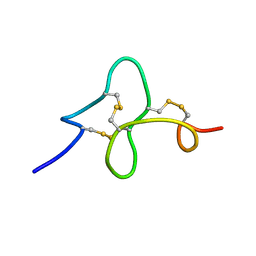 | | 24 NMR structures of AcAMP2-Like Peptide with Phenylalanine 18 mutated to Tryptophan | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZWU
 
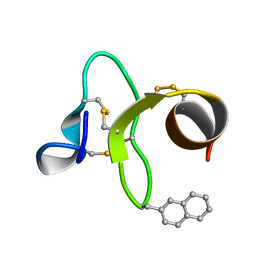 | | 30 NMR structures of AcAMP2-like peptide with non natural beta-(2-naphthyl)-alanine residue. | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 (ACMP2) | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues.
Chemistry, 11, 2005
|
|
1TEW
 
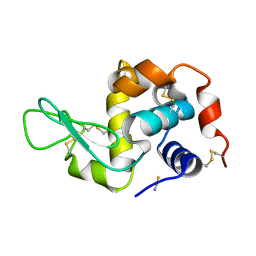 | |
1XFT
 
 | |
