6YVU
 
 | | Condensin complex from S.cerevisiae ATP-free apo non-engaged state | | Descriptor: | Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2,Smc2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Aragon, L, Haering, C.H, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YVD
 
 | | Head segment of the S.cerevisiae condensin holocomplex in presence of ATP | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Structural maintenance of chromosomes protein 2, ... | | Authors: | Merkel, F, Haering, C.H, Hassler, M, Lee, B.G, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YVV
 
 | | Condensin complex from S.cerevisiae ATP-free apo bridged state | | Descriptor: | Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Haering, C.H, Aragon, L, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6QJ2
 
 | |
6QJ4
 
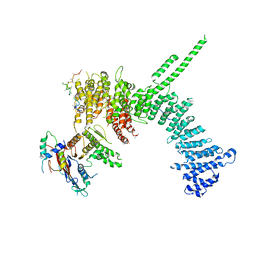 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex bound to the Smc4 ATPase head in complex with the C-terminal domain of Brn1 | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6QJ0
 
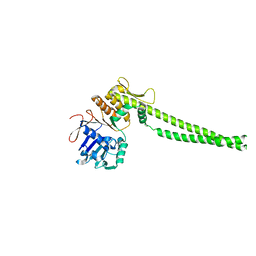 | |
6QJ1
 
 | |
6QJ3
 
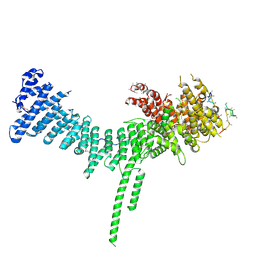 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2 | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6Q6E
 
 | | Structural and functional insights into the condensin ATPase cycle | | Descriptor: | Condensin complex subunit 2,Structural maintenance of chromosomes protein,Structural maintenance of chromosomes protein | | Authors: | Simon, B, Hassler, M, Haering, C.H, Hennig, J. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
2WD5
 
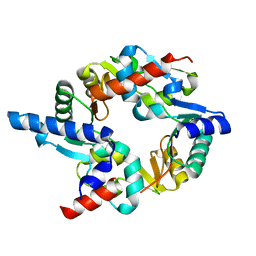 | | SMC hinge heterodimer (Mouse) | | Descriptor: | STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 1A, STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 3 | | Authors: | Michie, K.A, Haering, C.H, Nasmyth, K, Lowe, J. | | Deposit date: | 2009-03-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion.
Embo J., 30, 2011
|
|
5FRR
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5FRS
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN 1, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.073 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5FRP
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5OQR
 
 | |
5OQP
 
 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form I) | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, DNA (5'-D(*GP*AP*TP*GP*TP*GP*TP*AP*GP*CP*TP*AP*CP*AP*CP*AP*TP*C)-3'), ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQN
 
 | |
5OQQ
 
 | |
5OQO
 
 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Condensin complex subunit 2, Condensin complex subunit 3, ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
1W1W
 
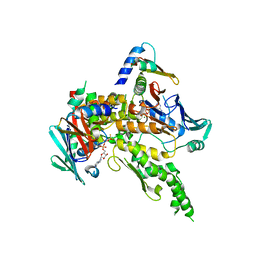 | | Sc Smc1hd:Scc1-C complex, ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SISTER CHROMATID COHESION PROTEIN 1, ... | | Authors: | Haering, C, Nasmyth, K, Lowe, J. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and stability of cohesin's Smc1-kleisin interaction.
Mol. Cell, 15, 2004
|
|
7QEN
 
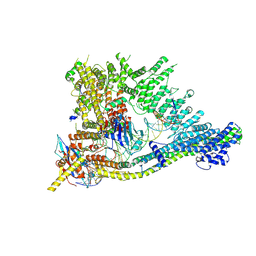 | | S.c. Condensin core in DNA- and ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
7QFW
 
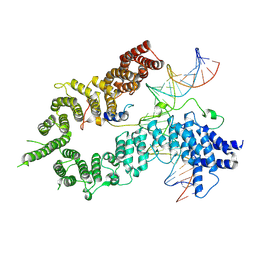 | | S.c. Condensin peripheral Ycg1 subcomplex bound to DNA | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Synthetic DNA ligand, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
6H8Q
 
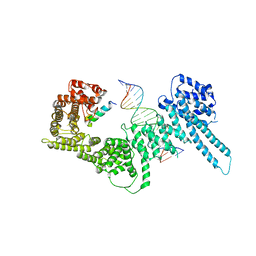 | | Structural basis for Scc3-dependent cohesin recruitment to chromatin | | Descriptor: | Cohesin subunit SCC3, DNA (5'-D(P*CP*TP*TP*TP*CP*GP*TP*TP*TP*CP*CP*TP*TP*GP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*CP*AP*AP*GP*GP*AP*AP*AP*CP*GP*AP*AP*AP*G)-3'), ... | | Authors: | Li, Y, Muir, K, Panne, D. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.631 Å) | | Cite: | Structural basis for Scc3-dependent cohesin recruitment to chromatin.
Elife, 7, 2018
|
|
