4HYJ
 
 | | Crystal structure of Exiguobacterium sibiricum rhodopsin | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Gushchin, I, Chervakov, P, Kuzmichev, P, Popov, A, Round, E, Borshchevskiy, V, Dolgikh, D, Kirpichnikov, M, Petrovskaya, L, Chupin, V, Arseniev, A, Gordeliy, V. | | Deposit date: | 2012-11-13 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3QDC
 
 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the active state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-18 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
3QAP
 
 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the ground state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
4XTN
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 4.9 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTO
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTL
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric blue form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, SODIUM ION, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5IJI
 
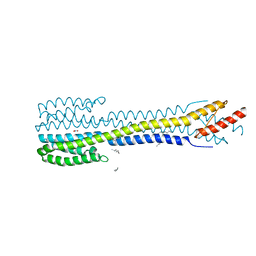 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in symmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-03-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
5JEF
 
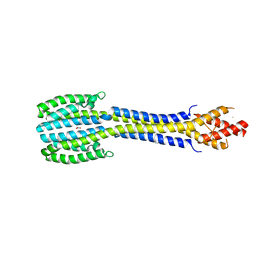 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in asymmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
5JEQ
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50K) in symmetric apo state | | Descriptor: | Nitrate/nitrite sensor protein NarQ, PHOSPHATE ION | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
6XYN
 
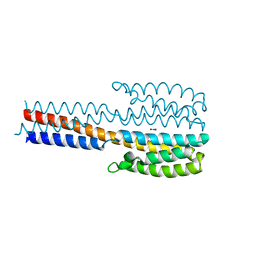 | | Crystal structure of a proteolytic fragment of NarQ comprising sensor and TM domains | | Descriptor: | NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Gordeliy, V. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a proteolytic fragment of the sensor histidine kinase NarQ
Crystals, 2020
|
|
6YUE
 
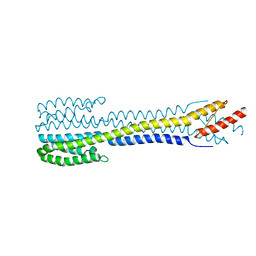 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50S variant) | | Descriptor: | Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Yuzhakova, A, Gordeliy, V. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sensor Histidine Kinase NarQ Activates via Helical Rotation, Diagonal Scissoring, and Eventually Piston-Like Shifts.
Int J Mol Sci, 21, 2020
|
|
7OO9
 
 | |
7AVP
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O state | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
7AVN
 
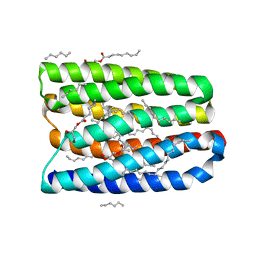 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the M-like state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
6SQG
 
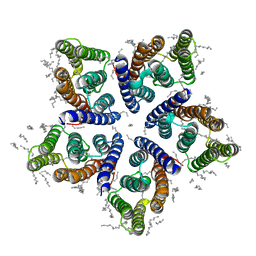 | | Crystal structure of viral rhodopsin OLPVRII | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Kovalev, K, Bratanov, D, Polovinkin, V, Astashkin, R, Popov, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique structure and function of viral rhodopsins.
Nat Commun, 10, 2019
|
|
7AVO
 
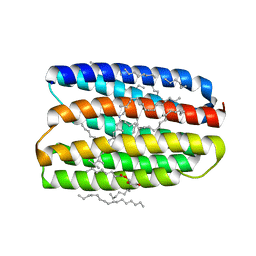 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1211 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
8CBB
 
 | |
4MD2
 
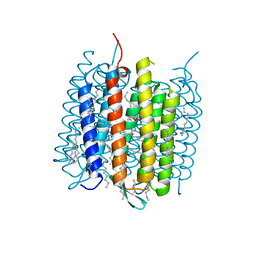 | | Ground state of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9VF8
 
 | | Structure of Meiothermus ruber Mrub_1259 LOV domain (MrLOV) | | Descriptor: | FLAVIN MONONUCLEOTIDE, histidine kinase | | Authors: | Semenov, O, Nazarenko, V, Yudenko, A, Remeeva, A, Borshchevskiy, V, Yang, Y, Gushchin, I. | | Deposit date: | 2025-06-10 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of Meiothermus ruber LOV domains
To Be Published
|
|
9VF7
 
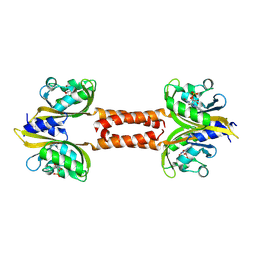 | | Structure of Meiothermus ruber Mrub_1259 LOV domain with N- and C-terminal alpha helices (MrLOVe) | | Descriptor: | FLAVIN MONONUCLEOTIDE, histidine kinase | | Authors: | Semenov, O, Nazarenko, V, Yudenko, A, Remeeva, A, Borshchevskiy, V, Yang, Y, Gushchin, I. | | Deposit date: | 2025-06-10 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Meiothermus ruber LOV domains
To Be Published
|
|
8PKY
 
 | |
8PM1
 
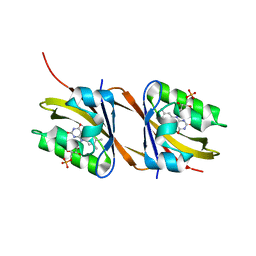 | |
9KME
 
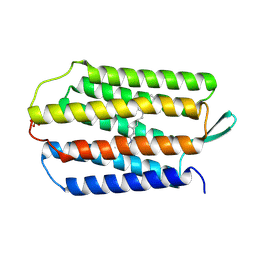 | | Crystal structure of soluble bacteriorhodopsin NeuroBR_A | | Descriptor: | RETINAL, SULFATE ION, soluble bacteriorhodopsin | | Authors: | Nikolaev, A, Remeeva, A, Kapranov, I, Borshchevskiy, V, Gushchin, I. | | Deposit date: | 2024-11-15 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Engineering of soluble bacteriorhodopsin.
Chem Sci, 16, 2025
|
|
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
4PXK
 
 | | Crystal structure of Haloarcula marismortui bacteriorhodopsin I D94N mutant | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Shevchenko, V, Gushchin, I, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2014-03-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli-Expressed Haloarcula marismortui Bacteriorhodopsin I in the Trimeric Form.
Plos One, 9, 2014
|
|
