1L5B
 
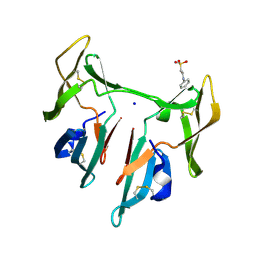 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | 著者 | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | 登録日 | 2002-03-06 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1L5E
 
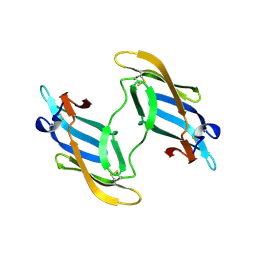 | | The domain-swapped dimer of CV-N in solution | | 分子名称: | Cyanovirin-N | | 著者 | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | 登録日 | 2002-03-06 | | 公開日 | 2002-06-05 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
2HIR
 
 | |
2NEF
 
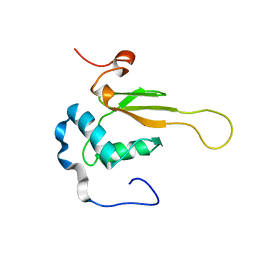 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | 分子名称: | NEGATIVE FACTOR (F-PROTEIN) | | 著者 | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | 登録日 | 1997-02-12 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
2EZZ
 
 | |
2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-07-25 | | 公開日 | 1997-12-03 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-07-25 | | 公開日 | 1997-12-03 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZG
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZF
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZK
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-10-04 | | 公開日 | 1998-01-14 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZL
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-10-04 | | 公開日 | 1998-01-14 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZM
 
 | |
2EZX
 
 | |
2EZY
 
 | |
2JZJ
 
 | | Structure of CrCVNH (C. richardii CVNH) | | 分子名称: | Cyanovirin-N homolog | | 著者 | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | 登録日 | 2008-01-09 | | 公開日 | 2008-03-25 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZL
 
 | | Structure of NcCVNH (N. CRASSA CVNH) | | 分子名称: | Cyanovirin-N homolog | | 著者 | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | 登録日 | 2008-01-09 | | 公開日 | 2008-03-25 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZK
 
 | | Structure of TbCVNH (T. BORCHII CVNH) | | 分子名称: | Cyanovirin-N homolog | | 著者 | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | 登録日 | 2008-01-09 | | 公開日 | 2008-03-25 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
3SAK
 
 | |
7UW3
 
 | |
3GB1
 
 | |
5UPW
 
 | |
1QCE
 
 | |
3CI2
 
 | |
