1M65
 
 | | YCDX PROTEIN | | Descriptor: | Hypothetical protein ycdX, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-07-12 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M68
 
 | | YCDX PROTEIN, TRINUCLEAR ZINC SITE | | Descriptor: | Hypothetical protein ycdX, SULFATE ION, ZINC ION | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2002-07-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a
trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1N2A
 
 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1MTC
 
 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
1NIJ
 
 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NRK
 
 | |
1PGA
 
 | |
1PGB
 
 | |
9RSA
 
 | |
8RSA
 
 | |
8ABP
 
 | |
7ABP
 
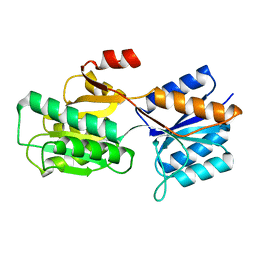 | |
5KNG
 
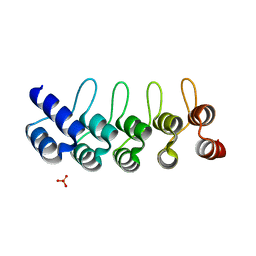 | | CRYSTAL STRUCTURE OF ANTI-IL-13 DARPIN 6G9 | | Descriptor: | DARPIN 6G9, GLYCEROL, PHOSPHATE ION | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
5KNH
 
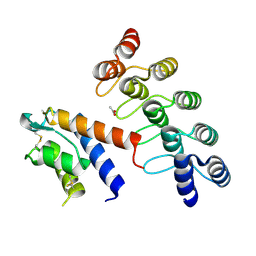 | | CRYSTAL STRUCTURE OF DARPIN 6G9 IN COMPLEX WITH CYNO IL-13 | | Descriptor: | ACETATE ION, DARPIN 6G9, IL13 | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
9ABP
 
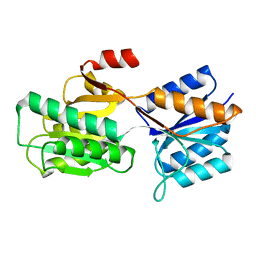 | |
6PTI
 
 | |
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
7JGU
 
 | | Structure of FN3tt mut | | Descriptor: | Fibronectin type-III domain-containing protein, PENTAETHYLENE GLYCOL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
5I1H
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1E
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV1-39 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I18
 
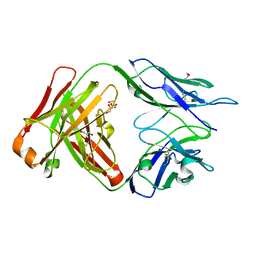 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1K
 
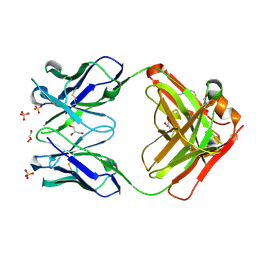 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV3-20 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I16
 
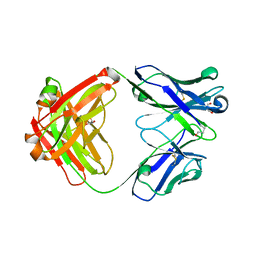 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV3-11 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
