5E6Y
 
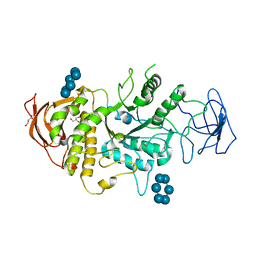 | | Crystal structure of E.Coli branching enzyme in complex with alpha cyclodextrin | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | Authors: | Feng, L, Nosrati, M, Geiger, J.H. | | Deposit date: | 2015-10-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5F58
 
 | |
5F6B
 
 | |
5FAZ
 
 | |
5FEN
 
 | |
5FFH
 
 | |
5F7G
 
 | |
3CWK
 
 | |
3D1J
 
 | |
3CX4
 
 | |
3COP
 
 | |
3D96
 
 | |
3D95
 
 | |
3D97
 
 | |
3FEL
 
 | |
3FEP
 
 | | Crystal structure of the R132K:R111L:L121E:R59W-CRABPII mutant complexed with a synthetic ligand (merocyanin) at 2.60 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
3FA9
 
 | |
3FA8
 
 | |
3FEN
 
 | |
3FA7
 
 | |
3FEK
 
 | |
2FRS
 
 | |
2FS6
 
 | |
2FR3
 
 | |
2FS7
 
 | |
