5XT5
 
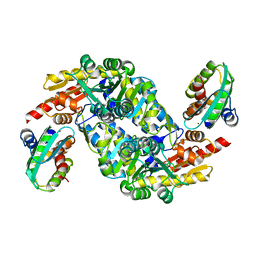 | | SufS-SufU complex from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, PYRIDOXAL-5'-PHOSPHATE, ZINC ION, ... | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
4YT4
 
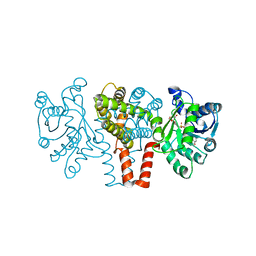 | |
4YT8
 
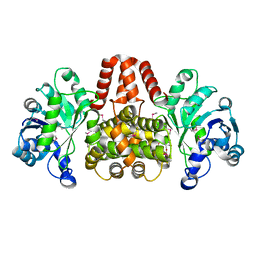 | | SeMet-labelled HmdII from Methanocaldococcus jannaschii | | Descriptor: | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4YT5
 
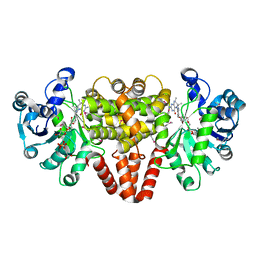 | | HmdII from Methanocaldococcus jannaschii with bound methylene-tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
4YTE
 
 | |
4YT2
 
 | | Hmd II from Methanocaldococcus jannaschii | | Descriptor: | DI(HYDROXYETHYL)ETHER, H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338, TRIETHYLENE GLYCOL | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards a functional identification of catalytically inactive [Fe]-hydrogenase paralogs.
Febs J., 282, 2015
|
|
5XT6
 
 | | A sulfur-transferring catalytic intermediate of SufS-SufU complex from Bacillus subtilis | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Cysteine desulfurase SufS, ZINC ION, ... | | Authors: | Fujishiro, T, Kunichika, K, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
6M2A
 
 | |
6M25
 
 | | Sirohydrochlorin nickelochelatase CfbA in P41 space group | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M27
 
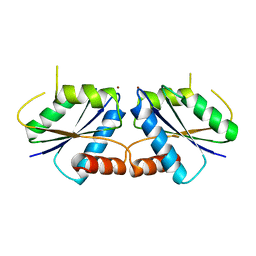 | | Sirohydrochlorin nickelochelatase CfbA in complex with Ni2+ | | Descriptor: | NICKEL (II) ION, Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M2E
 
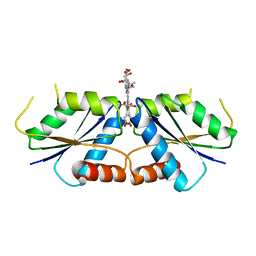 | | Sirohydrochlorin nickelochelatase CfbA in complex with sirohydrochlorin | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13-tetrahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Sirohydrochlorin cobaltochelatase | | Authors: | Fujishiro, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The nickel-sirohydrochlorin formation mechanism of the ancestral class II chelatase CfbA in coenzyme F430 biosynthesis.
Chem Sci, 12, 2021
|
|
6M2F
 
 | |
5D4U
 
 | | SAM-bound HcgC from Methanocaldococcus jannaschii | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5O
 
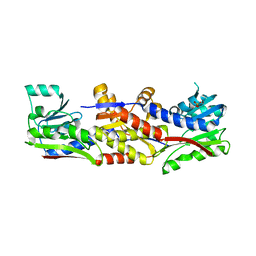 | | HcgC from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D4T
 
 | |
5D4V
 
 | | HcgC with SAH and a guanylylpyridinol (GP) derivative | | Descriptor: | 5'-O-[(R)-[(3,6-dimethyl-2-oxo-1,2-dihydropyridin-4-yl)oxy](hydroxy)phosphoryl]guanosine, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5T
 
 | |
5WT6
 
 | |
5WT5
 
 | | L-homocysteine-bound NifS from Helicobacter pylori | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshot of cysteine desulfurase NifS with L-cysteine in initiation of catalysis
to be published
|
|
7WSX
 
 | |
7E0L
 
 | |
3AWM
 
 | | Cytochrome P450SP alpha (CYP152B1) wild-type with palmitic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position
J.Biol.Chem., 286, 2011
|
|
3AWP
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant F288G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
3AWQ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant L78F | | Descriptor: | Fatty acid alpha-hydroxylase, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
7DE4
 
 | | Hybrid cluster protein (HCP) from Escherichia coli | | Descriptor: | FE-S-O HYBRID CLUSTER, Hydroxylamine reductase, IRON/SULFUR CLUSTER | | Authors: | Fujishiro, T, Ooi, M, Takaoka, K, Takahashi, Y. | | Deposit date: | 2020-11-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of Escherichia coli class II hybrid cluster protein, HCP, reveals a [4Fe-4S] cluster at the N-terminal protrusion.
Febs J., 288, 2021
|
|
