6C5Y
 
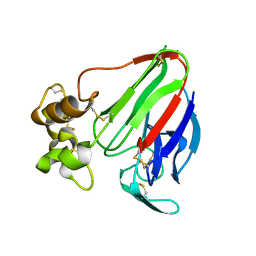 | | Crystal structure of thaumatin from microcrystals | | 分子名称: | Thaumatin-1 | | 著者 | Guo, G, Fuchs, M, Shi, W, Skinner, J, Berman, E, Ogata, C.M, Hendrickson, W.A, McSweeney, S, Liu, Q. | | 登録日 | 2018-01-17 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Sample manipulation and data assembly for robust microcrystal synchrotron crystallography.
IUCrJ, 5, 2018
|
|
4CJG
 
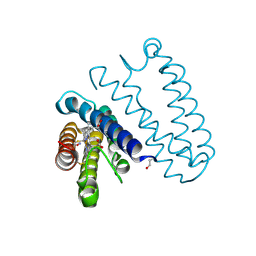 | | Spectroscopically validated structure of the 5 coordinate proximal NO adduct of cytochrome c prime from Alcaligenes xylosoxidans | | 分子名称: | CYTOCHROME C', HEME C, NITRIC OXIDE | | 著者 | Kekilli, D, Dworkowski, F, Fuchs, M, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-12-20 | | 公開日 | 2014-05-21 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6NCT
 
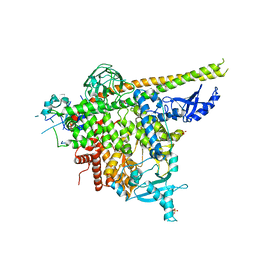 | | Structure of p110alpha/niSH2 - vector data collection | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | 著者 | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | 登録日 | 2018-12-12 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCK
 
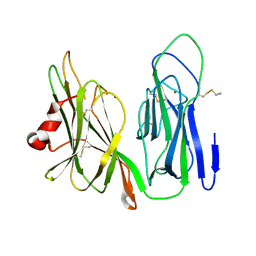 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | 分子名称: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | 著者 | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCI
 
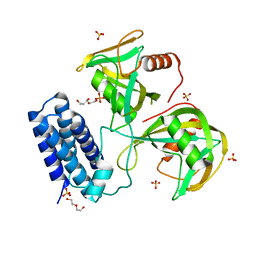 | | Crystal structure of CDP-Chase: Vector data collection | | 分子名称: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCH
 
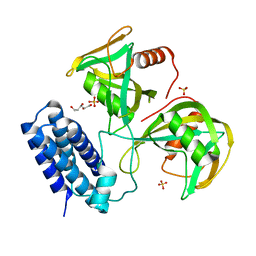 | | Crystal structure of CDP-Chase: Raster data collection | | 分子名称: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
8DCV
 
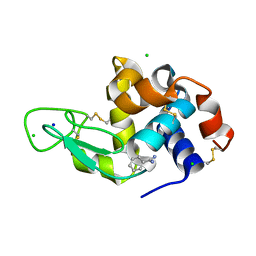 | | Lysozyme cluster 0043, NAG ligand | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | 登録日 | 2022-06-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCW
 
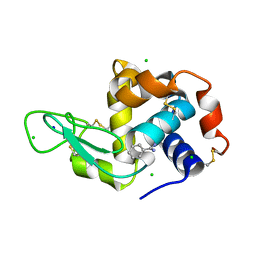 | | Lysozyme cluster 0062 (NAG and benzamidine ligands) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | 登録日 | 2022-06-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCU
 
 | | Lysozyme cluster 0028 (benzamidine ligand) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | 登録日 | 2022-06-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCT
 
 | | Lysozyme cluster 3 dual apo structure | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | 登録日 | 2022-06-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4CDA
 
 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | 分子名称: | CYTOCHROME C', HEME C, SULFATE ION | | 著者 | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-10-30 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CDV
 
 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 100K | | 分子名称: | CYTOCHROME C', HEME C, SULFATE ION | | 著者 | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-11-06 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CDY
 
 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 160K | | 分子名称: | CYTOCHROME C', HEME C | | 著者 | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-11-07 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CJO
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced at 180K using X-rays | | 分子名称: | CYTOCHROME C', HEME C | | 著者 | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-12-21 | | 公開日 | 2014-05-21 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CIP
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced using ascorbate | | 分子名称: | ASCORBIC ACID, CYTOCHROME C', HEME C, ... | | 著者 | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | 登録日 | 2013-12-13 | | 公開日 | 2014-05-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G51
 
 | | Crystallographic analysis of the interaction of nitric oxide with hemoglobin from Trematomus bernacchii in the T quaternary structure (fully ligated state). | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRIC OXIDE, ... | | 著者 | Merlino, A, Balsamo, A, Pica, A, Mazzarella, L, Vergara, A. | | 登録日 | 2012-07-17 | | 公開日 | 2013-01-16 | | 最終更新日 | 2019-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selective X-ray-induced NO photodissociation in haemoglobin crystals: evidence from a Raman-assisted crystallographic study.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TGA
 
 | |
3TGB
 
 | |
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.77 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | 分子名称: | NAD(+) hydrolase AbTIR | | 著者 | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | 登録日 | 2023-02-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
5RSO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000000226 | | 分子名称: | Non-structural protein 3, PARA ACETAMIDO BENZOIC ACID | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
