1QTP
 
 | |
1RJZ
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and Herpies Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RJY
 
 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Glycoprotein B Peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RK1
 
 | | Mhc Class I Natural H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1RK0
 
 | | Mhc Class I H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Glycoprotein B peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
1JTZ
 
 | | CRYSTAL STRUCTURE OF TRANCE/RANKL CYTOKINE. | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 11 | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity
J.Clin.Invest., 108, 2001
|
|
1KYD
 
 | |
1KY6
 
 | |
1KYF
 
 | |
1KY7
 
 | |
1KYU
 
 | |
1ML0
 
 | |
1MKF
 
 | |
1PSH
 
 | |
5VQ5
 
 | | Crystal Structure of the Lectin Domain From the F17-like Adhesin, UclD | | Descriptor: | Adhesin, IODIDE ION | | Authors: | Klein, R.D, Spaulding, C.N, Dodson, K.W, Pinkner, J.S, Hultgren, S.J, Fremont, D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist.
Nature, 546, 2017
|
|
6CRJ
 
 | |
5NWP
 
 | |
3NI0
 
 | | Crystal Structure of Mouse BST-2/Tetherin Ectodomain | | Descriptor: | Bone marrow stromal antigen 2, ISOPROPYL ALCOHOL | | Authors: | Scheaffer, S.M, Brett, T.J. | | Deposit date: | 2010-06-14 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biophysical Analysis of BST-2/Tetherin Ectodomains Reveals an Evolutionary Conserved Design to Inhibit Virus Release.
J.Biol.Chem., 286, 2011
|
|
2R6P
 
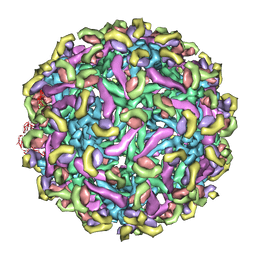 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6P4L
 
 | |
6P4J
 
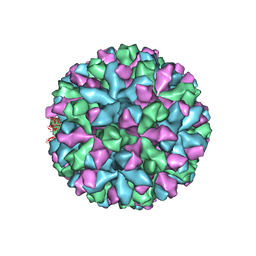 | | Mouse norovirus complexed with GCDCA | | Descriptor: | Capsid protein, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Smith, T.J, Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
6P4K
 
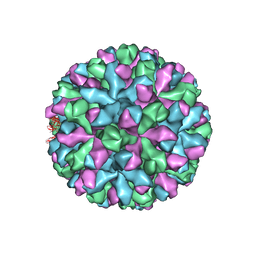 | | mouse norovirus complexed with TCA | | Descriptor: | Capsid protein, TAUROCHOLIC ACID | | Authors: | Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
1TYP
 
 | |
1FO0
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
