1ZZ3
 
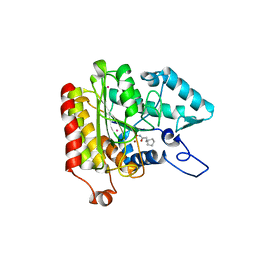 | | Crystal structure of a HDAC-like protein with CypX bound | | Descriptor: | 3-CYCLOPENTYL-N-HYDROXYPROPANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
1ZZ1
 
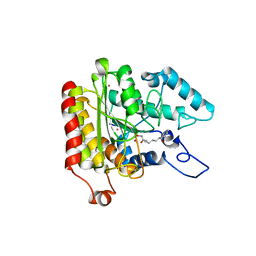 | | Crystal structure of a HDAC-like protein with SAHA bound | | Descriptor: | Histone deacetylase-like amidohydrolase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
2B2H
 
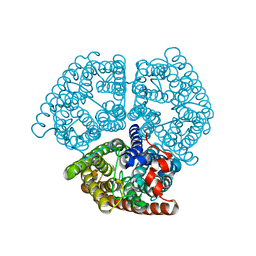 | |
2B2F
 
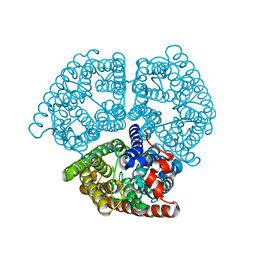 | |
2B2I
 
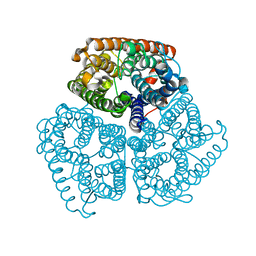 | |
2B2J
 
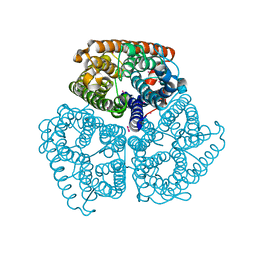 | | Ammonium Transporter Amt-1 from A. fulgidus (Xe) | | Descriptor: | XENON, ammonium transporter | | Authors: | Andrade, S.L.A, Dickmanns, A, Ficner, R, Einsle, O. | | Deposit date: | 2005-09-19 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B5D
 
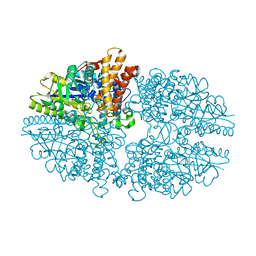 | | Crystal structure of the novel alpha-amylase AmyC from Thermotoga maritima | | Descriptor: | alpha-Amylase | | Authors: | Dickmanns, A, Ballschmiter, M, Liebl, W, Ficner, R. | | Deposit date: | 2005-09-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the novel alpha-amylase AmyC from Thermotoga maritima.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3EWI
 
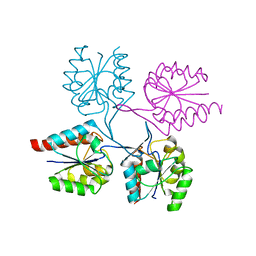 | | Structural analysis of the C-terminal domain of murine CMP-Sialic acid Synthetase | | Descriptor: | N-acylneuraminate cytidylyltransferase | | Authors: | Oschlies, M, Dickmanns, A, Stummeyer, K, Gerardy-Schahn, R, Ficner, R, Muenster-Kuehnel, A.K. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A C-terminal phosphatase module conserved in vertebrate CMP-sialic acid synthetases provides a tetramerization interface for the physiologically active enzyme.
J.Mol.Biol., 393, 2009
|
|
3GVJ
 
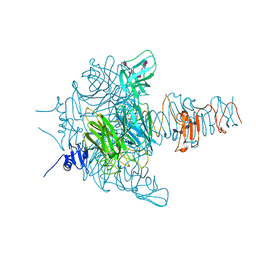 | | Crystal structure of an endo-neuraminidaseNF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GUD
 
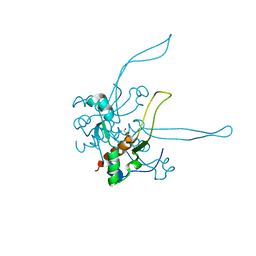 | | Crystal structure of a novel intramolecular chaperon | | Descriptor: | BROMIDE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GVL
 
 | | Crystal Structure of endo-neuraminidaseNF | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GW6
 
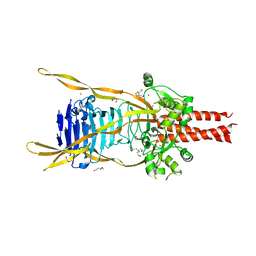 | | Intramolecular Chaperone | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GVK
 
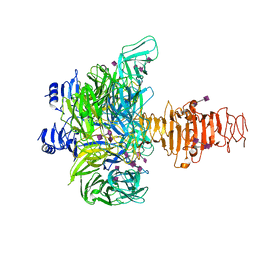 | | Crystal structure of endo-neuraminidase NF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
4KR9
 
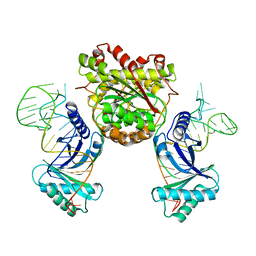 | |
4N6R
 
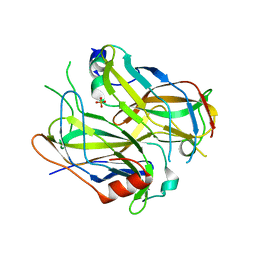 | | Crystal structure of VosA-VelB-complex | | Descriptor: | SULFATE ION, VelB, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4TMZ
 
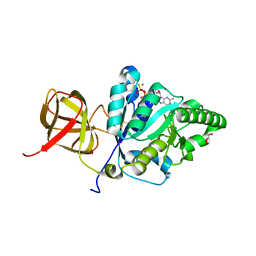 | |
4TN1
 
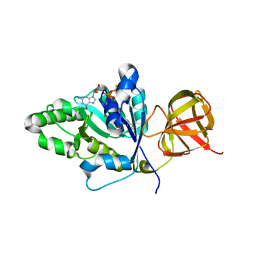 | |
4RV7
 
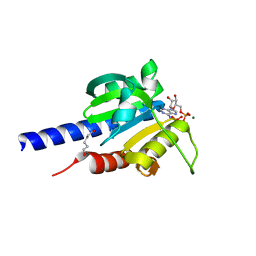 | | Characterization of an essential diadenylate cyclase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Diadenylate cyclase, HEXANE-1,6-DIOL, ... | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Analysis of the Essential Diadenylate Cyclase CdaA from Listeria monocytogenes.
J.Biol.Chem., 290, 2015
|
|
4TMW
 
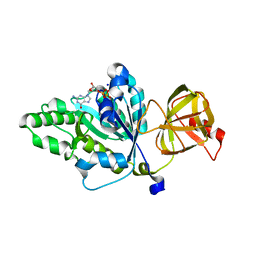 | |
4RLE
 
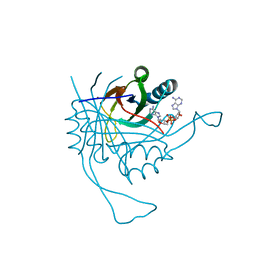 | | Crystal structure of the c-di-AMP binding PII-like protein DarA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, NICKEL (II) ION, Uncharacterized protein YaaQ | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2014-10-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification, Characterization, and Structure Analysis of the Cyclic di-AMP-binding PII-like Signal Transduction Protein DarA.
J.Biol.Chem., 290, 2015
|
|
4TMV
 
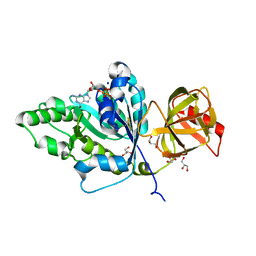 | |
4TMT
 
 | | Translation initiation factor eIF5B (517-858) mutant D533A from C. thermophilum, bound to GTPgammaS | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
4TMX
 
 | | Translation initiation factor eIF5B (517-858) mutant D533N from C. thermophilum, bound to GTP and sodium | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
4JQ9
 
 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
4K51
 
 | | Crystal Structure of the PCI domain of eIF3a | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2013-04-12 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes.
Nucleic Acids Res., 42, 2014
|
|
