3HKF
 
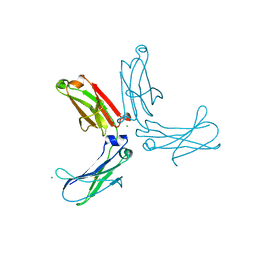 | | Murine unglycosylated IgG Fc fragment | | Descriptor: | CHLORIDE ION, Igh protein, MAGNESIUM ION | | Authors: | Feige, M.J, Nath, S, Catharino, S.R, Weinfurtner, D, Steinbacher, S, Buchner, J. | | Deposit date: | 2009-05-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the murine unglycosylated IgG1 Fc fragment
J.Mol.Biol., 391, 2009
|
|
4Q9B
 
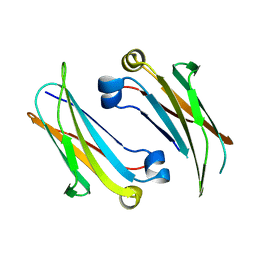 | | IgNAR antibody domain C2 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q9C
 
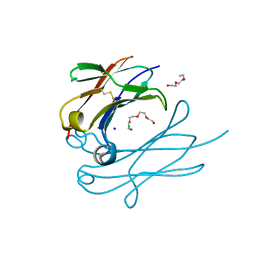 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q97
 
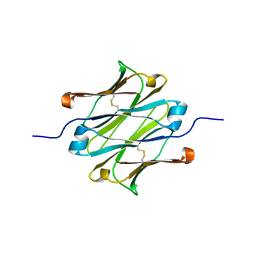 | | IgNAR antibody domain C1 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MKL
 
 | |
6ZMD
 
 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
8CIL
 
 | |
