6JSU
 
 | |
6JST
 
 | |
4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | 著者 | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-08-07 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4ZO5
 
 | | PDE10 complexed with 4-isopropoxy-2-(2-(3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl)ethyl)isoindoline-1,3-dione | | 分子名称: | 2-{2-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}-4-(propan-2-yloxy)-1H-isoindole-1,3(2H)-dione, 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, MAGNESIUM ION, ... | | 著者 | Yan, Y. | | 登録日 | 2015-05-06 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of [(11)C]MK-8193 as a PET tracer to measure target engagement of phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7DJJ
 
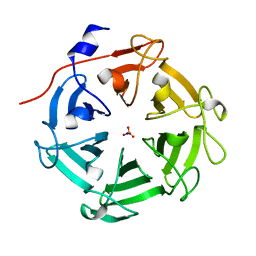 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | 分子名称: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.69806433 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
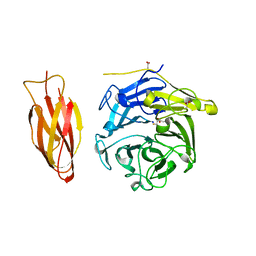 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.70000112 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
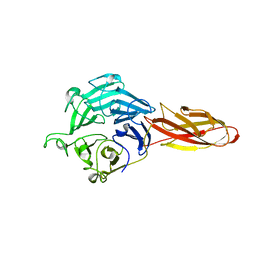 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.96077824 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
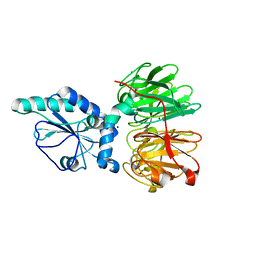 | | Structure of four truncated and mutated forms of quenching protein | | 分子名称: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | 著者 | Yu, G.M, Pan, X.W, Li, M. | | 登録日 | 2020-11-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.80145121 Å) | | 主引用文献 | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
