6SKU
 
 | | Legionella effector AnkX in complex with human Rab1b | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphocholine transferase AnkX, ... | | Authors: | Ernst, S, Ecker, F, Kaspers, M, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Legionellaeffector AnkX displaces the switch II region for Rab1b phosphocholination.
Sci Adv, 6, 2020
|
|
6FHO
 
 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa - new refinement | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable FAD-dependent monooxygenase | | Authors: | Belviso, B.D, Drees, S.L, Ernst, S, Jagmann, N, Hennecke, U, Fetzner, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PqsL uses reduced flavin to produce 2-hydroxylaminobenzoylacetate, a preferred PqsBC substrate in alkyl quinolone biosynthesis inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
1EE9
 
 | | CRYSTAL STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH NAD | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-31 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
1EDZ
 
 | | STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-28 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
8ANP
 
 | | Legionella effector Lem3 mutant D190A in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Phosphocholine hydrolase Lem3, SULFATE ION, ... | | Authors: | Kaspers, M.S, Pogenberg, V, Ernst, S, Ecker, F, Pett, C, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
1RTC
 
 | | THE STRUCTURE OF RECOMBINANT RICIN A CHAIN AT 2.3 ANGSTROMS | | Descriptor: | RICIN | | Authors: | Mlsna, D, Monzingo, A.F, Katzin, B.J, Ernst, S, Robertus, J.D. | | Deposit date: | 1992-10-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of recombinant ricin A chain at 2.3 A.
Protein Sci., 2, 1993
|
|
1DU5
 
 | | THE CRYSTAL STRUCTURE OF ZEAMATIN. | | Descriptor: | ZEAMATIN | | Authors: | Batalia, M.A, Monzingo, A.F, Ernst, S, Roberts, W, Robertus, J.D. | | Deposit date: | 2000-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family.
Nat.Struct.Biol., 3, 1996
|
|
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
1IBV
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE BOUND WITH HISTIDINE METHYL ESTER AT-170 C | | Descriptor: | HISTIDINE DECARBOXYLASE BETA CHAIN, HISTIDINE-METHYL-ESTER, Histidine decarboxylase alpha chain | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1IBW
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE BOUND WITH HISTIDINE METHYL ESTER AT 25 C | | Descriptor: | HISTIDINE DECARBOXYLASE BETA CHAIN, HISTIDINE-METHYL-ESTER, Histidine decarboxylase alpha chain | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1IBU
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE AT 25 C | | Descriptor: | HISTIDINE DECARBOXYLASE ALPHA CHAIN, HISTIDINE DECARBOXYLASE BETA CHAIN | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1HQ6
 
 | | STRUCTURE OF PYRUVOYL-DEPENDENT HISTIDINE DECARBOXYLASE AT PH 8 | | Descriptor: | HISTIDINE DECARBOXYLASE | | Authors: | Schelp, E, Worley, S, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | pH-induced structural changes regulate histidine decarboxylase activity in Lactobacillus 30a.
J.Mol.Biol., 306, 2001
|
|
1IBT
 
 | | STRUCTURE OF THE D53,54N MUTANT OF HISTIDINE DECARBOXYLASE AT-170 C | | Descriptor: | HISTIDINE DECARBOXYLASE ALPHA CHAIN, HISTIDINE DECARBOXYLASE BETA CHAIN | | Authors: | Worley, S, Schelp, E, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Proteins, 46, 2002
|
|
1LL4
 
 | | STRUCTURE OF C. IMMITIS CHITINASE 1 COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE 1 | | Authors: | Bortone, K, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2002-04-26 | | Release date: | 2002-09-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE STRUCTURE OF AN ALLOSAMIDIN COMPLEX WITH THE Coccidioides IMMITIS CHITINASE DEFINES A ROLE FOR A SECOND ACID RESIDUE IN SUBSTRATE-ASSISTED MECHANISM
J.Mol.Biol., 320, 2002
|
|
1LL7
 
 | |
1LL6
 
 | |
6SW2
 
 | |
6SW1
 
 | | Crystal Structure of P. aeruginosa PqsL: R41Y, I43R, G45R, C105G mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mattevi, A, Rovida, S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer.
Nat Commun, 11, 2020
|
|
2AAI
 
 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
3MF7
 
 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
1OBT
 
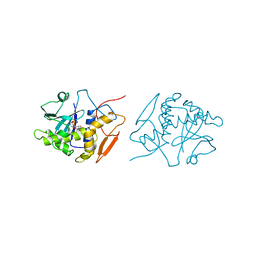 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OBS
 
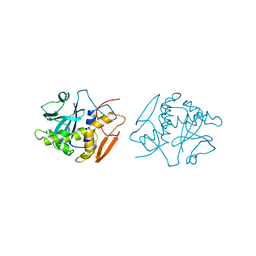 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
3MF8
 
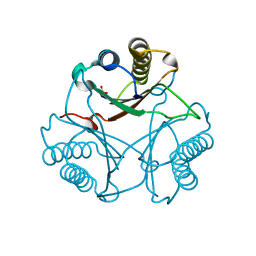 | | Crystal Structure of Native cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
1D2K
 
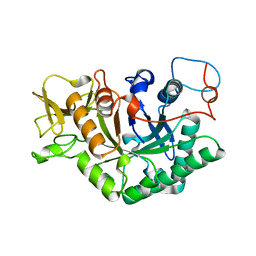 | | C. IMMITIS CHITINASE 1 AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE 1 | | Authors: | Hollis, T, Monzingo, A.F, Bortone, K, Ernst, S.R, Cox, R, Robertus, J.D. | | Deposit date: | 1999-09-23 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of a chitinase from the pathogenic fungus Coccidioides immitis.
Protein Sci., 9, 2000
|
|
4MAD
 
 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
