3KSK
 
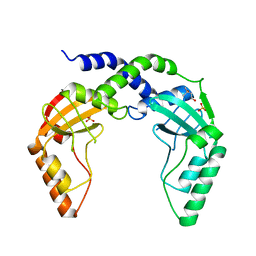 | | Crystal Structure of single chain PvuII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | Meramveliotaki, C, Hountas, A, Eliopoulos, E, Kokkinidis, M. | | Deposit date: | 2009-11-23 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of single chain PvuII
To be Published
|
|
4V33
 
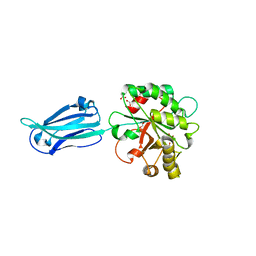 | | Crystal structure of the putative polysaccharide deacetylase BA0330 from bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSACCHARIDE DEACETYLASE-LIKE PROTEIN, ... | | Authors: | Giastas, P, Andreou, A, Bouriotis, V, Eliopoulos, E. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Two Putative Polysaccharide Deacetylases are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus Anthracis.
J.Biol.Chem., 290, 2015
|
|
1FN4
 
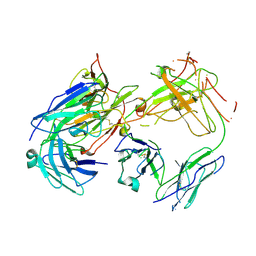 | | CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES | | Descriptor: | MONOCLONAL ANTIBODY AGAINST ACETYLCHOLINE RECEPTOR | | Authors: | Poulas, K, Eliopoulos, E, Vatzaki, E, Navaza, J, Kontou, M. | | Deposit date: | 2000-08-21 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Fab198, an efficient protector of the acetylcholine receptor against myasthenogenic antibodies.
Eur.J.Biochem., 268, 2001
|
|
3N7H
 
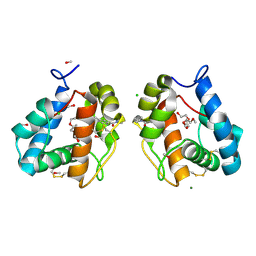 | | Crystal structure of Odorant Binding Protein 1 from Anopheles gambiae (AgamOBP1) with DEET (N,N-Diethyl-meta-toluamide) and PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2010-05-27 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anopheles gambiae odorant binding protein crystal complex with the synthetic repellent DEET: implications for structure-based design of novel mosquito repellents.
Cell.Mol.Life Sci., 69, 2012
|
|
4IJ7
 
 | | Crystal structure of Odorant Binding Protein 48 from Anopheles gambiae (AgamOBP48) with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Odorant binding protein-8, SODIUM ION | | Authors: | Zographos, S.E, Tsitsanou, K.E, Drakou, C.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and Solution Studies of the "Plus-C" Odorant-binding Protein 48 from Anopheles gambiae: CONTROL OF BINDING SPECIFICITY THROUGH THREE-DIMENSIONAL DOMAIN SWAPPING.
J.Biol.Chem., 288, 2013
|
|
4KYN
 
 | |
6GE7
 
 | |
6GHH
 
 | |
6GFS
 
 | |
6GF9
 
 | |
