7R4W
 
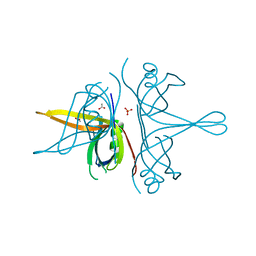 | | Single stranded DNA binding protein SSB M5 from Fervidobacterium gondwanense | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Single-stranded DNA-binding protein | | Authors: | Hakansson, M, Svensson, L.A, Werbowy, O, Al-Karadaghi, S, Kaczorowski, T, Kaczorowska, A.K, Dorawa, S. | | Deposit date: | 2022-02-09 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a single stranded DNA binding protein from Fervidobacterium gondwanense
To Be Published
|
|
7B3N
 
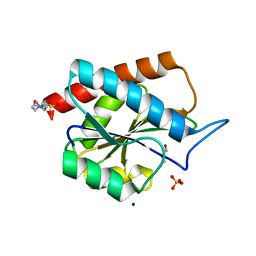 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
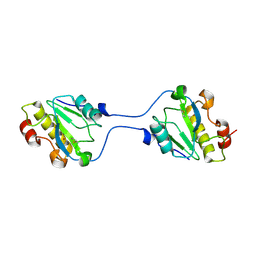
6FHG
 
 | |
