6ENO
 
 | | Double cubane cluster oxidoreductase | | Descriptor: | CITRIC ACID, Dehydratase family protein, Double cubane cluster, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | ATP-dependent substrate reduction at an [Fe8S9] double-cubane cluster.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ELQ
 
 | | Carbon Monoxide Dehydrogenase IV from Carboxydothermus hydrogenoformans | | Descriptor: | Carbon Monoxide Dehydrogenase, FE(4)-NI(1)-S(5) CLUSTER with Oxygen, IRON/SULFUR CLUSTER | | Authors: | Domnik, L, Goetzl, S, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2017-09-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | CODH-IV: A High-Efficiency CO-Scavenging CO Dehydrogenase with Resistance to O2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8CAV
 
 | | Discovery of the lanthipeptide Curvocidin and structural insights into its trifunctional synthetase CuvL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CuvA, MAGNESIUM ION, ... | | Authors: | Sigurdsson, A, Martins, B.M, Duettmann, S.A, Jasyk, M, Dimos-Roehl, B, Schoepf, F, Gemannter, M, Knittel, C.H, Schnegotyzki, R, Schmid, B, Kosol, S, Gonzalez-Viegas, M, Seidel, M, Huegelland, M, Leimkuehler, S, Dobbek, H, Mainz, A, Suessmuth, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4EHU
 
 | |
4EIA
 
 | |
4EHT
 
 | |
4EO0
 
 | | crystal structure of the pilus binding domain of the filamentous phage IKe | | Descriptor: | Attachment protein G3P | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
4EMI
 
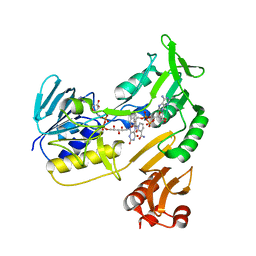 | | Toluene dioxygenase reductase in reduced state in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TodA | | Authors: | Lin, T.Y, Werther, T, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Suppression of Electron Transfer to Dioxygen by Charge Transfer and Electron Transfer Complexes in the FAD-dependent Reductase Component of Toluene Dioxygenase.
J.Biol.Chem., 287, 2012
|
|
4EO1
 
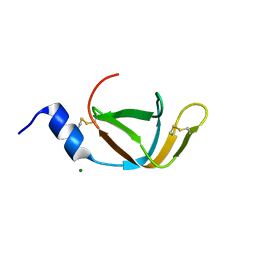 | | crystal structure of the TolA binding domain from the filamentous phage IKe | | Descriptor: | Attachment protein G3P, MAGNESIUM ION | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
4EMJ
 
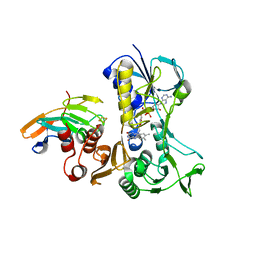 | | Complex between the reductase and ferredoxin components of toluene dioxygenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, TodA, ... | | Authors: | Lin, T.Y, Werther, T, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Suppression of Electron Transfer to Dioxygen by Charge Transfer and Electron Transfer Complexes in the FAD-dependent Reductase Component of Toluene Dioxygenase.
J.Biol.Chem., 287, 2012
|
|
1RU3
 
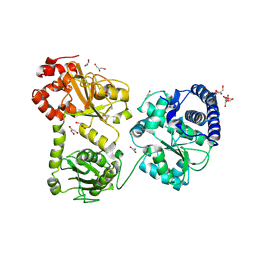 | | Crystal Structure of the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans | | Descriptor: | Acetyl-CoA synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Svetlitchnyi, V, Dobbek, H, Meyer-Klaucke, W, Meins, T, Thiele, B, Rmer, P, Huber, R, Meyer, O. | | Deposit date: | 2003-12-11 | | Release date: | 2003-12-23 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T3Q
 
 | | Crystal structure of quinoline 2-Oxidoreductase from Pseudomonas Putida 86 | | Descriptor: | DIOXOSULFIDOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonin, I, Martins, B.M, Purvanov, V, Fetzner, S, Huber, R, Dobbek, H. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
STRUCTURE, 12, 2004
|
|
1U8V
 
 | | Crystal Structure of 4-Hydroxybutyryl-CoA Dehydratase from Clostridium aminobutyricum: Radical catalysis involving a [4Fe-4S] cluster and flavin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gamma-aminobutyrate metabolism dehydratase/isomerase, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Dobbek, H, Cinkaya, I, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-08-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 4-hydroxybutyryl-CoA dehydratase: radical catalysis involving a [4Fe-4S] cluster and flavin.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5FLI
 
 | | enzyme-substrate complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NICKEL (II) ION, QUERCETINASE QUED | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-On O2-Ni Complex.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5FLE
 
 | | High resolution NI,FE-CODH-320 mV with CN state | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ciaccafava, A, Tombolelli, D, Domnik, L, Fesseler, J, Jeoung, J.-H, Dobbek, H, Mroginski, M.A, Hildebrandt, P, Zebger, I. | | Deposit date: | 2015-10-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | When the inhibitor tells more than the substrate: the cyanide-bound state of a carbon monoxide dehydrogenase.
Chem Sci, 7, 2016
|
|
5FLH
 
 | | Free state of Ni-quercetinase | | Descriptor: | DIMETHYL SULFOXIDE, NICKEL (II) ION, NITRATE ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5FLJ
 
 | | enzyme-substrate-dioxygen complex of Ni-quercetinase | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, NICKEL (II) ION, ... | | Authors: | Jeoung, J.-H, Nianios, D, Fetzner, S, Dobbek, H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-on O2 -Ni Complex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5I7P
 
 | | Crystal structure of Fkbp12-IF(SlyD), a chimeric protein of human Fkbp12 and the insert in flap domain of Ecoli SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,FKBP-type peptidyl-prolyl cis-trans isomerase SlyD,Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Jakob, R.P, Knappe, T.A, Dobbek, H, Schmid, F.X. | | Deposit date: | 2016-02-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural and Functional Analysis of Chaperone Domain Insertion in Fkbp12
To Be Published
|
|
5I7Q
 
 | |
7O0D
 
 | |
7NYP
 
 | | monomeric acetyl-CoA synthase in closed conformation | | Descriptor: | 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CO-methylating acetyl-CoA synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7NZ5
 
 | | monomeric acetyl-CoA synthase with Zn at the proximal position of cluster A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CITRIC ACID, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7NYS
 
 | | monomeric acetyl-CoA synthase in closed conformation with carbon monoxide bound to the Ni proximal of cluster A | | Descriptor: | $l^{3}-oxidanylidynemethylnickel, 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
