5D8C
 
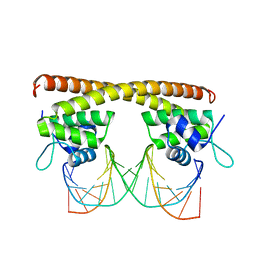 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to promoter DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*AP*AP*CP*TP*CP*TP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*TP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3'), MerR family regulator protein | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
5E01
 
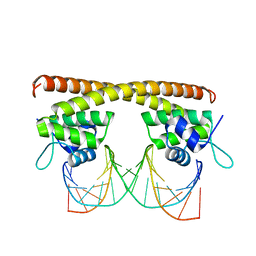 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
7RGV
 
 | |
4B6G
 
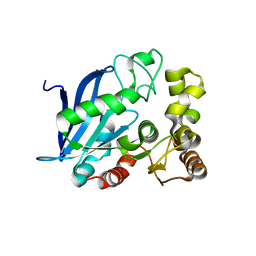 | | The Crystal Structure of the Neisserial Esterase D. | | Descriptor: | PUTATIVE ESTERASE | | Authors: | Counago, R.M, Kobe, B. | | Deposit date: | 2012-08-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms.
Antioxid. Redox Signal., 18, 2013
|
|
8C46
 
 | |
5D90
 
 | |
