1QUS
 
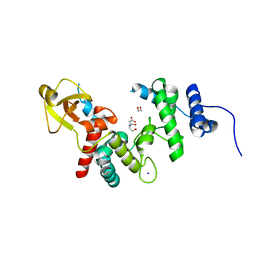 | | 1.7 A RESOLUTION STRUCTURE OF THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, BICINE, LYTIC MUREIN TRANSGLYCOSYLASE B, ... | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
1QUT
 
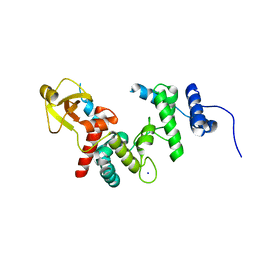 | | THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI IN COMPLEX WITH N-ACETYLGLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LYTIC MUREIN TRANSGLYCOSYLASE B, SODIUM ION | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
3H8F
 
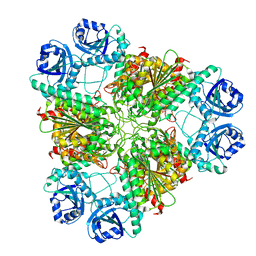 | | High pH native structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | BICARBONATE ION, Cytosol aminopeptidase, MANGANESE (II) ION, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3H8E
 
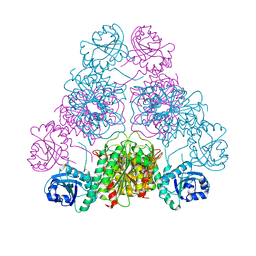 | |
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3HZ3
 
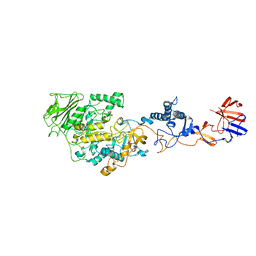 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180(D1025N)-sucrose complex | | Descriptor: | CALCIUM ION, Glucansucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-06-23 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PYG
 
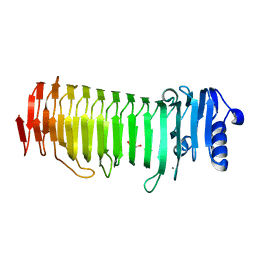 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachmann, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
2PYH
 
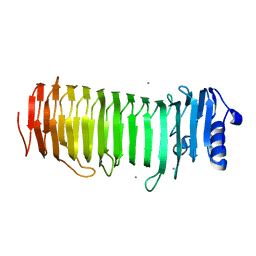 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide | | Descriptor: | CALCIUM ION, Poly(beta-D-mannuronate) C5 epimerase 4, alpha-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachman, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
2Y9W
 
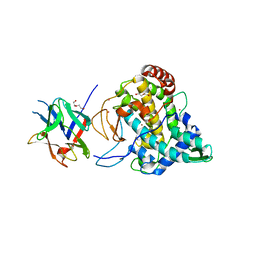 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2YFT
 
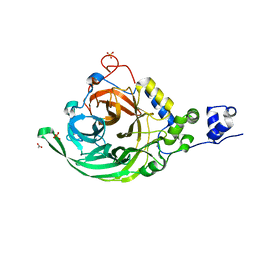 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with 1-kestose | | Descriptor: | ACETATE ION, CALCIUM ION, LEVANSUCRASE, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2YII
 
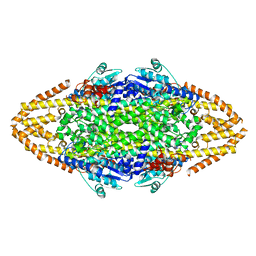 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2YFS
 
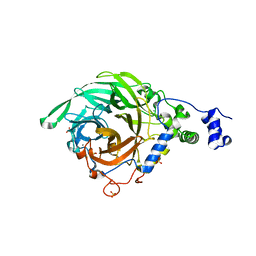 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with sucrose | | Descriptor: | CALCIUM ION, LEVANSUCRASE, SULFATE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2Y9X
 
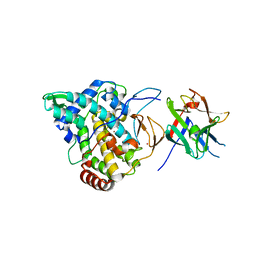 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit, with inhibitor tropolone | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, COPPER (II) ION, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2YKV
 
 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, BETA-TRANSAMINASE | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
2YKU
 
 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, BETA-TRANSAMINASE, GLYCEROL, ... | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
2YFR
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2YKY
 
 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | (3S)-3-amino-3-phenylpropanoic acid, 1,2-ETHANEDIOL, BENZENE, ... | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
2YKX
 
 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BETA-TRANSAMINASE, ... | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
2WYC
 
 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ in complex with 3-oxo-lauric acid | | Descriptor: | 3-OXODODECANOIC ACID, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, ... | | Authors: | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2X2J
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2X2I
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2WYB
 
 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ with a covalently bound dodecanoic acid | | Descriptor: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL, ... | | Authors: | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WYD
 
 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ in complex with dodecanoic acid | | Descriptor: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL, ... | | Authors: | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2X1C
 
 | | The crystal structure of precursor acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | ACYL-COENZYME, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
2X2H
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
