1EO5
 
 | |
1HDE
 
 | |
1CGX
 
 | |
1CGW
 
 | |
1CGY
 
 | |
1CGV
 
 | |
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
1UKR
 
 | | STRUCTURE OF ENDO-1,4-BETA-XYLANASE C | | Descriptor: | ENDO-1,4-B-XYLANASE I | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1996-08-23 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
J.Mol.Biol., 263, 1996
|
|
2GDC
 
 | | Structure of Vinculin VD1 / IpaA560-633 complex | | Descriptor: | Invasin ipaA, Vinculin | | Authors: | Hamiaux, C, van Eerde, A, Parsot, C, Broos, J, Dijkstra, B.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri.
Embo Rep., 7, 2006
|
|
1CRU
 
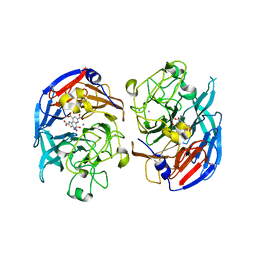 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2GVY
 
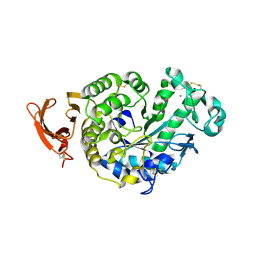 | |
2HTN
 
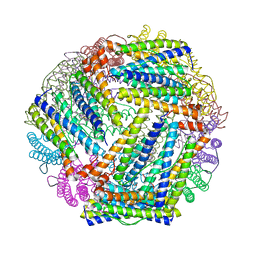 | | E. coli bacterioferritin in its as-isolated form | | Descriptor: | Bacterioferritin, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | van Eerde, A, Wolterink-Van Loo, S, Van Der Oost, J, Dijkstra, B.W. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fortuitous structure determination of 'as-isolated' Escherichia coli bacterioferritin in a novel crystal form.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2HIH
 
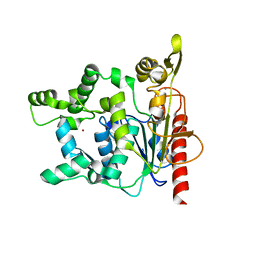 | | Crystal structure of Staphylococcus hyicus lipase | | Descriptor: | CALCIUM ION, Lipase 46 kDa form, ZINC ION | | Authors: | Tiesinga, J.J.W, van Pouderoyen, G, Nardini, M, Dijkstra, B.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of phospholipase activity of Staphylococcus hyicus lipase.
J.Mol.Biol., 371, 2007
|
|
1DTU
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE: A MUTANT Y89D/S146P COMPLEXED TO AN HEXASACCHARIDE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production.
J.Mol.Biol., 296, 2000
|
|
4C3Y
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
4C3X
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
1FW3
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PHOSPHOLIPASE A | | Authors: | Snijder, H.J, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-06-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli.
J.Mol.Biol., 309, 2001
|
|
1FW2
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | CALCIUM ION, OUTER MEMBRANE PHOSPHOLIPASE A | | Authors: | Snijder, H.J, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-06-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli.
J.Mol.Biol., 309, 2001
|
|
2DHE
 
 | |
2DHD
 
 | |
2DHC
 
 | |
2EDC
 
 | |
2EDA
 
 | |
1A47
 
 | | CGTASE FROM THERMOANAEROBACTERIUM THERMOSULFURIGENES EM1 IN COMPLEX WITH A MALTOHEXAOSE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1998-02-11 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1.
J.Biol.Chem., 273, 1998
|
|
1CQ1
 
 | | Soluble Quinoprotein Glucose Dehydrogenase from Acinetobacter Calcoaceticus in Complex with PQQH2 and Glucose | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
