1BU4
 
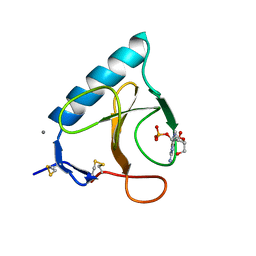 | | RIBONUCLEASE 1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
4A3X
 
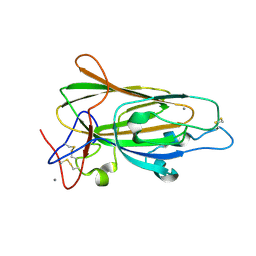 | | Structure of the N-terminal domain of the Epa1 adhesin (Epa1-Np) from the pathogenic yeast Candida glabrata, in complex with calcium and lactose | | Descriptor: | CALCIUM ION, EPA1P, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ielasi, F.S, Decanniere, K, Willaert, R.G. | | Deposit date: | 2011-10-05 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Epithelial Adhesin 1 (Epa1P) from the Human-Pathogenic Yeast Candida Glabrata : Structural and Functional Study of the Carbohydrate-Binding Domain
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4BU4
 
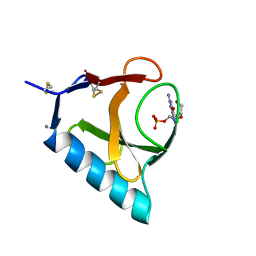 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
2BU4
 
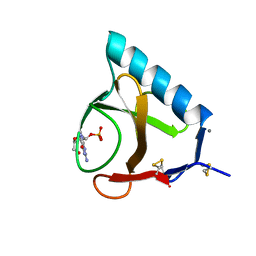 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
1G6V
 
 | | Complex of the camelid heavy-chain antibody fragment CAB-CA05 with bovine carbonic anhydrase | | Descriptor: | ANTIBODY HEAVY CHAIN, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Desmyter, A, Decanniere, K, Muyldermans, S, Wyns, L. | | Deposit date: | 2000-11-08 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody.
J.Biol.Chem., 276, 2001
|
|
1HOZ
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX | | Descriptor: | CALCIUM ION, GLYCEROL, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1AJK
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJO
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1YC8
 
 | | cAbAn33- Y37V/E44G/R45L triple mutant | | Descriptor: | anti-VSG immunoglobulin heavy chain variable domain cAbAn33 | | Authors: | Conrath, K, Vincke, C, Stijlemans, B, Schymkowitz, J, Wyns, L, Muyldermans, S, Loris, R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antigen Binding and Solubility Effects upon the Veneering of a Camel VHH in Framework-2 to Mimic a VH.
J.Mol.Biol., 350, 2005
|
|
1YC7
 
 | | cAbAn33 VHH fragment against VSG | | Descriptor: | SULFATE ION, anti-VSG immunoglobulin heavy chain variable domain cAbAn33 | | Authors: | Conrath, K, Vincke, C, Stijlemans, B, Schymkowitz, J, Wyns, L, Muyldermans, S, Loris, R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antigen Binding and Solubility Effects upon the Veneering of a Camel VHH in Framework-2 to Mimic a VH.
J.Mol.Biol., 350, 2005
|
|
1Q8F
 
 | |
1YOE
 
 | |
