6I1W
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Gp(PSU)pC-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*GP*AP*CP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*GP*(PSU)P*CP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
6I1V
 
 | | Structure of the RNA duplex containing pseudouridine residue (5'-Cp(PSU)pG-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*CP*AP*GP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*CP*(PSU)P*GP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
8G8Z
 
 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | Authors: | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G2W
 
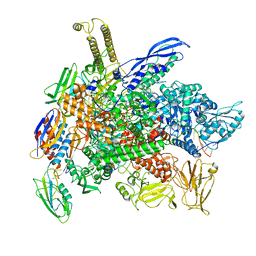 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4W
 
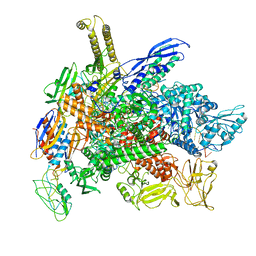 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G1S
 
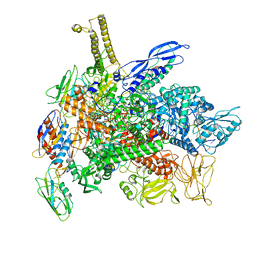 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G00
 
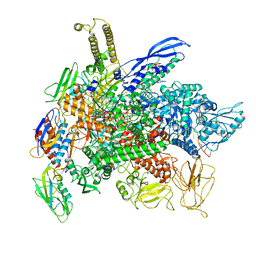 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-mer), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G7E
 
 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5DE1
 
 | | Crystal structure of human IDH1 in complex with GSK321A | | Descriptor: | (7R)-1-(4-fluorobenzyl)-N-{3-[(1S)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrol-2-ylcarbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Concha, N.O, Smallwood, A, Qi, H. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New IDH1 mutant inhibitors for treatment of acute myeloid leukemia.
Nat.Chem.Biol., 11, 2015
|
|
