1P3Q
 
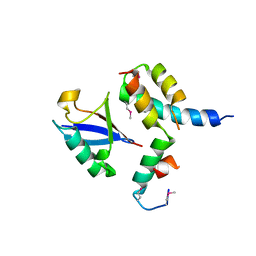 | | Mechanism of Ubiquitin Recognition by the CUE Domain of VPS9 | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E.A, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2003-04-18 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p.
Cell(Cambridge,Mass.), 113, 2003
|
|
1MN3
 
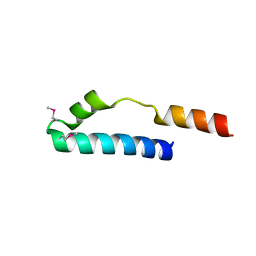 | | Cue domain of yeast Vps9p | | Descriptor: | Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2002-09-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p
Cell(Cambridge,Mass.), 113, 2003
|
|
3TW1
 
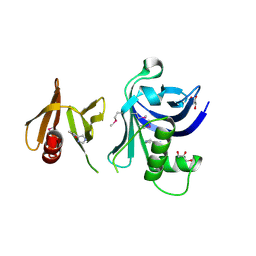 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
3TVV
 
 | |
2LH0
 
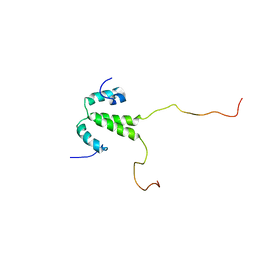 | |
3GGZ
 
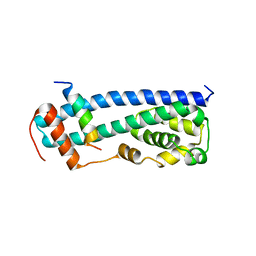 | |
3FSS
 
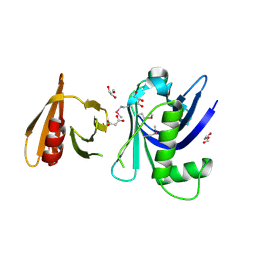 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
3GGY
 
 | | Crystal Structure of S.cerevisiae Ist1 N-terminal domain | | Descriptor: | Increased sodium tolerance protein 1 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2009-03-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis.
MOLECULAR BIOLOGY OF THE CELL, 20, 2009
|
|
2RKK
 
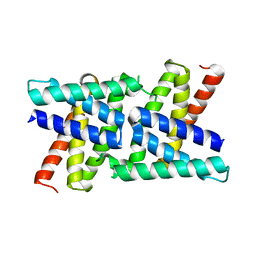 | |
2RKL
 
 | | Crystal Structure of S.cerevisiae Vta1 C-terminal domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Xiao, J, Xia, H, Zhou, J, Xu, Z. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of vta1 function in the multivesicular body sorting pathway.
Dev.Cell, 14, 2008
|
|
5LOZ
 
 | | STRUCTURE OF YEAST ENT1 ENTH DOMAIN | | Descriptor: | Epsin-1 | | Authors: | Tanner, N, Prag, G. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A bacterial genetic selection system for ubiquitylation cascade discovery.
Nat.Methods, 13, 2016
|
|
