8BPR
 
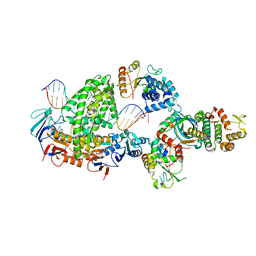 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | Descriptor: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9FRX
 
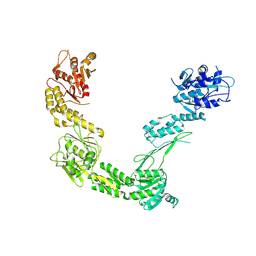 | | Porcine Retinol-Binding Protein 3 (RBP3) | | Descriptor: | Retinol binding protein 3 | | Authors: | Kaushik, V, Gessa, L, Kumar, N, Pinkas, M, Czarnocki-Cieciura, M, Palczewski, K, Novacek, J, Fernandes, H. | | Deposit date: | 2024-06-19 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | CryoEM structure and small-angle X-ray scattering analyses of porcine retinol-binding protein 3.
Open Biology, 15, 2025
|
|
8BDC
 
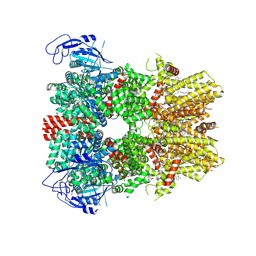 | | Human apo TRPM8 in a closed state (composite map) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Palchevskyi, S, Czarnocki-Cieciura, M, Vistoli, G, Gervasoni, S, Nowak, E, Beccari, A.R, Nowotny, M, Talarico, C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of human TRPM8 channel.
Commun Biol, 6, 2023
|
|
7R06
 
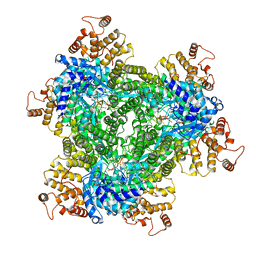 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Figiel, M, Nowotny, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R07
 
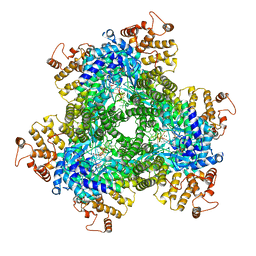 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), MAGNESIUM ION | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | Descriptor: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | Authors: | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
7O24
 
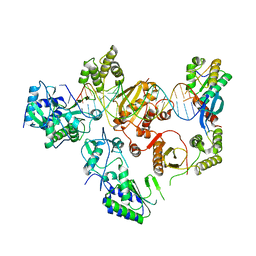 | |
7Z0Z
 
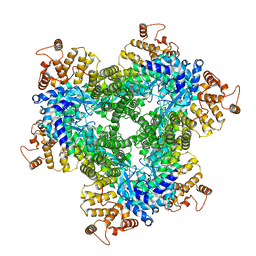 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis, Y44F variant | | Descriptor: | AbiK | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
8A93
 
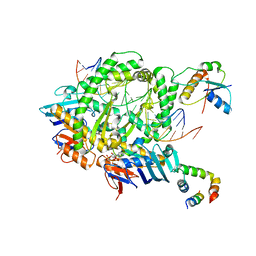 | | Complex of RecF-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A8J
 
 | | Complex of RecF and DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AB0
 
 | | Complex of RecO-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA repair protein RecO, Oligo1, Oligo2, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8P00
 
 | | Cryo-EM structure of Rotavirus B NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Chamera, S, Nowotny, M. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of rotavirus B NSP2 reveals its unique tertiary architecture.
J.Virol., 98, 2024
|
|
7R08
 
 | | Abortive infection DNA polymerase Abi-P2 | | Descriptor: | Reverse transcriptase | | Authors: | Gapinska, M.A, Figiel, M, Czarnocki Cieciura, M, Nowotny, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
6F3H
 
 | | Crystal structure of Dss1 exoribonuclease active site mutant D477N from Candida glabrata | | Descriptor: | Exoribonuclease II, mitochondrial, MAGNESIUM ION, ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
6F4A
 
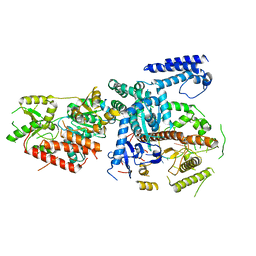 | | Yeast mitochondrial RNA degradosome complex mtEXO | | Descriptor: | Exoribonuclease II, mitochondrial, RNA (5'-R(P*AP*GP*AP*UP*AP*C)-3'), ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
7O0H
 
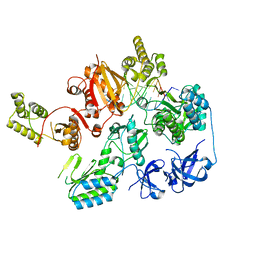 | |
7O0G
 
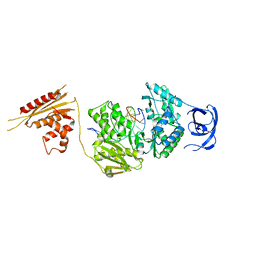 | |
4CT5
 
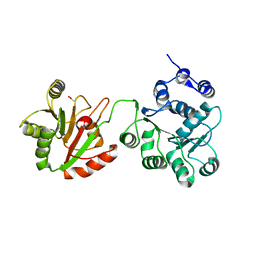 | | DDX6 | | Descriptor: | ACETATE ION, DDX6 | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4CT6
 
 | | CNOT9-CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4CV5
 
 | | yeast NOT1 CN9BD-CAF40 complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, HEXATANTALUM DODECABROMIDE, PROTEIN CAF40 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4CT7
 
 | | CNOT9-CNOT1 complex with bound tryptophan | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, ... | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
4CT4
 
 | | CNOT1 MIF4G domain - DDX6 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ozgur, S, Basquin, J, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4- not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
