7ZBU
 
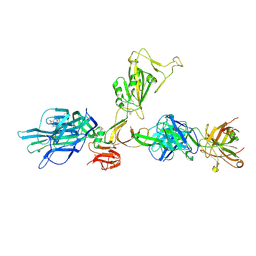 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
2XJA
 
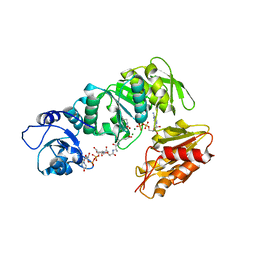 | | Structure of MurE from M.tuberculosis with dipeptide and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Basavannacharya, C, Moody, P.R, Bhakta, S, Keep, N. | | Deposit date: | 2010-07-03 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Essential Residues for the Enzyme Activity of ATP-Dependent Mure Ligase from Mycobacterium Tuberculosis.
Protein Cell, 1, 2010
|
|
6H3K
 
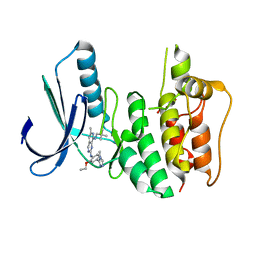 | | Introduction of a methyl group curbs metabolism of pyrido[3,4-d]pyrimidine MPS1 inhibitors and enables the discovery of the Phase 1 clinical candidate BOS172722. | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, ~{N}8-(2,2-dimethylpropyl)-~{N}2-[2-ethoxy-4-(4-methyl-1,2,4-triazol-3-yl)phenyl]-6-methyl-pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Woodward, H.L, Hoelder, S. | | Deposit date: | 2018-07-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Introduction of a Methyl Group Curbs Metabolism of Pyrido[3,4- d]pyrimidine Monopolar Spindle 1 (MPS1) Inhibitors and Enables the Discovery of the Phase 1 Clinical Candidate N2-(2-Ethoxy-4-(4-methyl-4 H-1,2,4-triazol-3-yl)phenyl)-6-methyl- N8-neopentylpyrido[3,4- d]pyrimidine-2,8-diamine (BOS172722).
J. Med. Chem., 61, 2018
|
|
8REC
 
 | |
8REE
 
 | |
8REB
 
 | |
8REA
 
 | |
8RED
 
 | |
8RE4
 
 | |
5DMA
 
 | |
5MKR
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in up-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, CITRATE ANION, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MKS
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in down-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2Y1Z
 
 | | Human alphaB Crystallin ACD R120G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-CRYSTALLIN B CHAIN | | Authors: | Clark, A.R, Bagneris, C, Naylor, C.E, Keep, N.H, Slingsby, C. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of R120G Disease Mutant of Human Alphab-Crystallin Domain Dimer Shows Closure of a Groove
J.Mol.Biol., 408, 2011
|
|
2Y1Y
 
 | | Human alphaB crystallin ACD(residues 71-157) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALPHA-CRYSTALLIN B CHAIN, | | Authors: | Naylor, C.E, Bagneris, C, Clark, A.R, Keep, N.H, Slingsby, C. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of R120G Disease Mutant of Human Alphab-Crystallin Domain Dimer Shows Closure of a Groove
J.Mol.Biol., 408, 2011
|
|
2Y22
 
 | | Human AlphaB-crystallin Domain (residues 67-157) | | Descriptor: | ALPHA-CRYSTALLIN B | | Authors: | Naylor, C.E, Bagneris, C, Clark, A.R, Keep, N.H, Slingsby, C. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of R120G Disease Mutant of Human Alphab-Crystallin Domain Dimer Shows Closure of a Groove
J.Mol.Biol., 408, 2011
|
|
