3RTY
 
 | | Structure of an Enclosed Dimer Formed by The Drosophila Period Protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Period circadian protein | | Authors: | King, H.A, Hoelz, A, Crane, B.R, Young, M.W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an enclosed dimer formed by the Drosophila period protein.
J.Mol.Biol., 413, 2011
|
|
3SFT
 
 | |
4HI4
 
 | |
4I44
 
 | |
4I3M
 
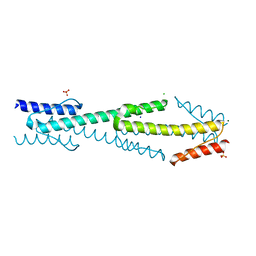 | | Aer2 poly-HAMP domains: L44H HAMP1 CW-lock mutant | | Descriptor: | Aerotaxis transducer Aer2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Airola, M.V, Sukomon, N, Crane, B.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HAMP Domain Conformers That Propagate Opposite Signals in Bacterial Chemoreceptors.
Plos Biol., 11, 2013
|
|
4HZ1
 
 | | Crystal Structure of Pseudomonas aeruginosa azurin with iron(II) at the copper-binding site. | | Descriptor: | ACETATE ION, Azurin, FE (II) ION | | Authors: | McLaughlin, M.P, Retegan, M, Bill, E, Payne, T.M, Shafaat, H.S, Pea, S, Sudhamsu, J, Ensign, A.A, Crane, B.R, Neese, F, Holland, P.L. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azurin as a Protein Scaffold for a Low-coordinate Nonheme Iron Site with a Small-molecule Binding Pocket.
J.Am.Chem.Soc., 134, 2012
|
|
4JPB
 
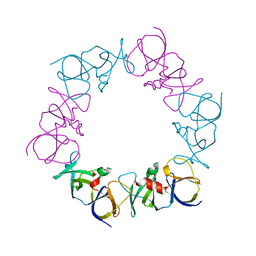 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
1I5B
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE | | Descriptor: | ACETATE ION, CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I53
 
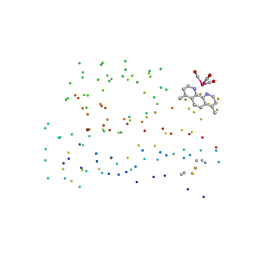 | | RE(I)-TRICARBONYL DIIMINE (Q107H)) AZURIN | | Descriptor: | 4,7-DIMETHYL-[1,10]PHENANTHROLINE, AZURIN, COPPER (II) ION, ... | | Authors: | Di Bilio, A.J, Crane, B.R, Wehbi, W.A, Kiser, C.N, Abu-Omar, M.M, Carlos, R.M, Richards, J.H, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-02-24 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Properties of photogenerated tryptophan and tyrosyl radicals in structurally characterized proteins containing rhenium(I) tricarbonyl diimines.
J.Am.Chem.Soc., 123, 2001
|
|
1I5C
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I59
 
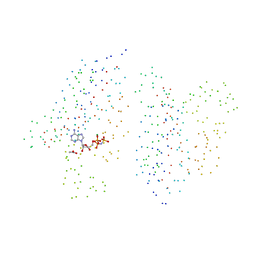 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ADPNP AND MAGENSIUM | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
2NOX
 
 | | Crystal structure of tryptophan 2,3-dioxygenase from Ralstonia metallidurans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Zhang, Y, Kang, S.A, Mukherjee, T, Bale, S, Crane, B.R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
Biochemistry, 46, 2007
|
|
2PD8
 
 | |
2PD7
 
 | |
2PDR
 
 | |
2A4M
 
 | | Structure of Trprs II bound to ATP | | Descriptor: | TRYPTOPHAN, Tryptophanyl-tRNA synthetase II | | Authors: | Buddha, M.R, Crane, B.R. | | Deposit date: | 2005-06-29 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Tryptophanyl-tRNA Synthetase II from Deinococcus radiodurans Bound to ATP and Tryptophan: Insight into subunit cooperativity and domain motions linked to catalysis
J.Biol.Chem., 280, 2005
|
|
2AN2
 
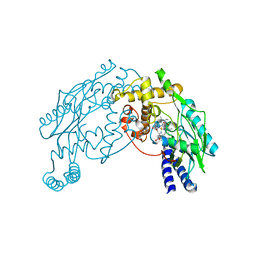 | | P332G, A333S Double mutant of the Bacillus subtilis Nitric Oxide Synthase | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, P332G A333S double mutant of Nitric Oxide Synthase from Bacillus subtilis, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a loose dimer: an intermediate in nitric oxide synthase assembly
J.Mol.Biol., 352, 2005
|
|
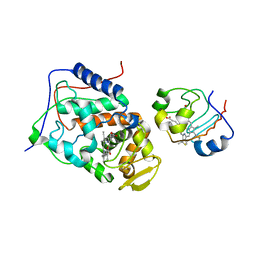
2B12
 
 | |
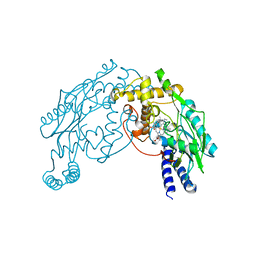
2AN0
 
 | |
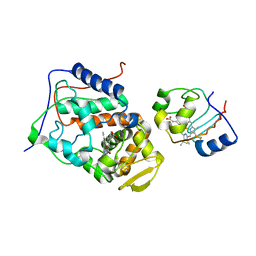
2B0Z
 
 | |
2AMO
 
 | |
2B10
 
 | |
2B11
 
 | |
2BCN
 
 | |
2CH7
 
 | |
