8P5C
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P86
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P57
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar X77. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P54
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar MG-132. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5B
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5A
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer R. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P56
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar X77. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P55
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar MG-132. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
5EUU
 
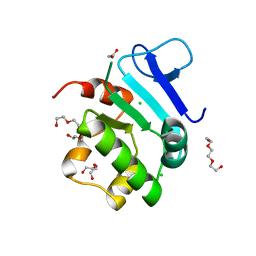 | | Rat prestin STAS domain in complex with chloride | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Prestin,Prestin, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUS
 
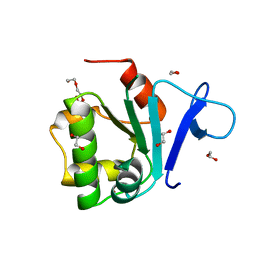 | | Rat prestin STAS domain in complex with bromide | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Prestin,Rat prestin STAS domain, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUW
 
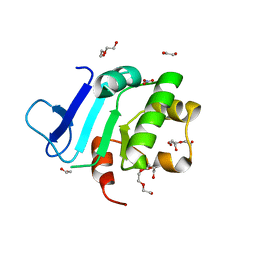 | | Rat prestin STAS domain in complex with nitrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NITRATE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUX
 
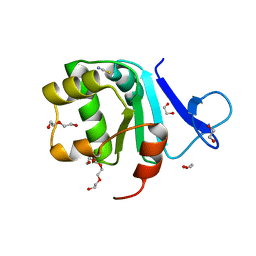 | | Rat prestin STAS domain in complex with thiocyanate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Prestin,Prestin, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUZ
 
 | | Rat prestin STAS domain in complex with iodide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EZB
 
 | | Chicken prestin STAS domain | | Descriptor: | Chicken prestin STAS domain,Chicken prestin STAS domain, GLYCEROL, OXALATE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
8QQG
 
 | | Structure of BRAF in Complex With Exarafenib (KIN-2787). | | Descriptor: | (3~{S})-~{N}-[4-methyl-3-[2-morpholin-4-yl-6-[[(2~{R})-1-oxidanylpropan-2-yl]amino]pyridin-4-yl]phenyl]-3-[2,2,2-tris(fluoranyl)ethyl]pyrrolidine-1-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Schmitt, A, Costanzi, E, Kania, R, Chen, Y.K. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.979 Å) | | Cite: | Structure of BRAF in Complex With Exarafenib (KIN-2787).
To Be Published
|
|
