7QOQ
 
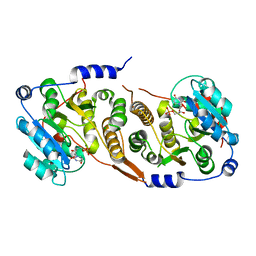 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S, Costa, D. | | Deposit date: | 2021-12-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium
To Be Published
|
|
7PHO
 
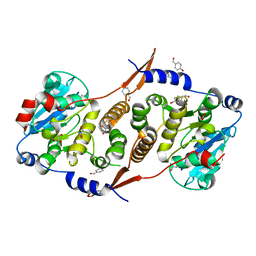 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde | | Descriptor: | BICARBONATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Nunes-Costa, D, Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-17 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde
To Be Published
|
|
7PVL
 
 | |
7QCP
 
 | |
7QIB
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP
To Be Published
|
|
7QI9
 
 | |
7P5L
 
 | |
7P8G
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MALONATE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-07-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form
To Be Published
|
|
7PD5
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid
To Be Published
|
|
7PDO
 
 | |
2W03
 
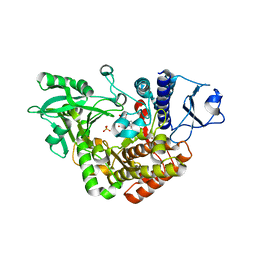 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with adenosine, sulfate and citrate from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE, CITRIC ACID, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W04
 
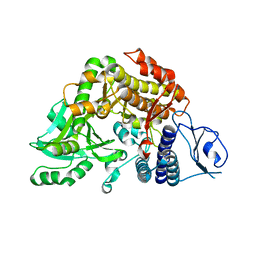 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2W02
 
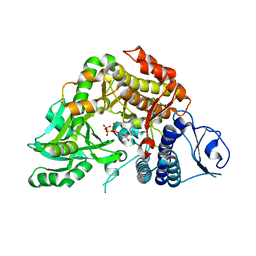 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with ATP from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
Nat. Chem. Biol., 5, 2009
|
|
5FDL
 
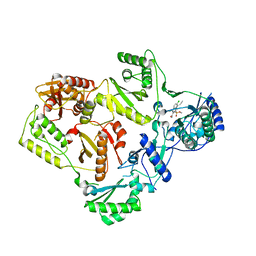 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5CZB
 
 | | HCV NS5B IN COMPLEX WITH LIGAND IDX17119-5 | | Descriptor: | 1-[4-(7-amino-5-methylpyrazolo[1,5-a]pyrimidin-2-yl)phenyl]-3-{[(R)-(2,4-dimethylphenyl)(methoxy)phosphoryl]amino}-1H-pyrazole-4-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pierra, C, Dousson, C, Augustin, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2LIF
 
 | | Solution Structure of KKGF | | Descriptor: | Core protein p21 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2011-08-29 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of hepatitis C virus core-e1 signal Peptide and requirements for cleavage of the genotype 3a signal sequence by signal Peptide peptidase.
J.Virol., 86, 2012
|
|
6HWK
 
 | | Glucosamine kinase (crystal form B) | | Descriptor: | CHLORIDE ION, Glucosamine kinase, MAGNESIUM ION | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.688 Å) | | Cite: | Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
Mbio, 10, 2019
|
|
6HWL
 
 | | Glucosamine kinase in complex with glucosamine, ADP and inorganic phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
Mbio, 10, 2019
|
|
6HWJ
 
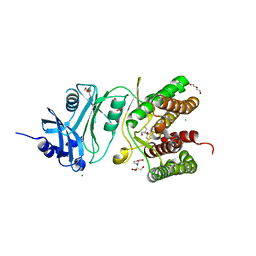 | | Glucosamine kinase (crystal form A) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
Mbio, 10, 2019
|
|
7PE4
 
 | |
4V84
 
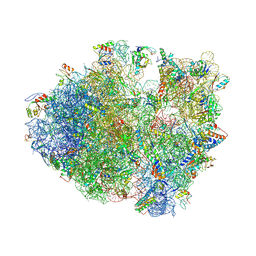 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V83
 
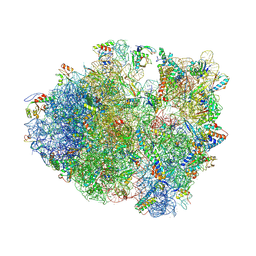 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
3TO3
 
 | | Crystal Structure of Petrobactin Biosynthesis Protein AsbB from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kim, Y, Eschenfeldt, W, Stols, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-03 | | Release date: | 2011-10-05 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Functional and Structural Analysis of the Siderophore Synthetase AsbB through Reconstitution of the Petrobactin Biosynthetic Pathway from Bacillus anthracis.
J.Biol.Chem., 287, 2012
|
|
2HQ6
 
 | | Structure of the Cyclophilin_CeCYP16-Like Domain of the Serologically Defined Colon Cancer Antigen 10 from Homo Sapiens | | Descriptor: | GLYCEROL, IODIDE ION, Serologically defined colon cancer antigen 10 | | Authors: | Walker, J.R, Davis, T, Paramanathan, R, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
