7AMF
 
 | |
4QXX
 
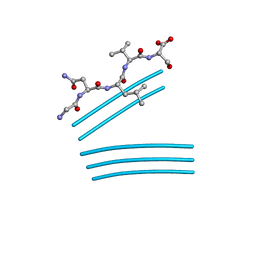 | |
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
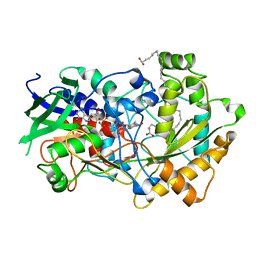
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
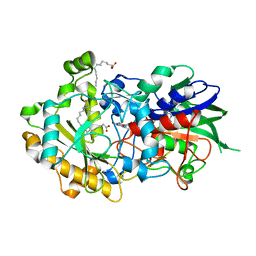
6YRZ
 
 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
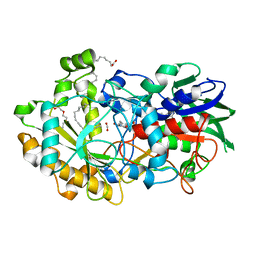
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8C34
 
 | |
8C33
 
 | |
8C36
 
 | |
8C37
 
 | |
8C32
 
 | |
8C31
 
 | |
8C35
 
 | |
4A16
 
 | | Structure of mouse Acetylcholinesterase complex with Huprine derivative | | Descriptor: | (1-{4-[(7S,11S)-12-AMINO-3-CHLORO-6,7,10,11-TETRAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-9-YL]BUTYL}-1H-1,2,3-TRIAZOL-4-YL)METHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Carletti, E, Colletier, J.P, Nachon, F, Weik, M, Ronco, C, Jean, L, Renard, P.Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Huprine Derivatives as Sub-Nanomolar Human Acetylcholinesterase Inhibitors: From Rational Design to Validation by X-Ray Crystallography.
Chemmedchem, 7, 2012
|
|
8BKH
 
 | |
8BKN
 
 | |
4C69
 
 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6R6V
 
 | | Structure of recombinant human butyrylcholinesterase in complex with a fluorescent coumarin-based probe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Nachon, F, Knez, D, Gobec, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of potent reversible selective inhibitors of butyrylcholinesterase as fluorescent probes.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6R6W
 
 | | Structure of recombinant human butyrylcholinesterase in complex with a fluorescent NBD-based probe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Nachon, F, Knez, D, Gobec, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Development of potent reversible selective inhibitors of butyrylcholinesterase as fluorescent probes.
J Enzyme Inhib Med Chem, 35, 2020
|
|
5DTX
 
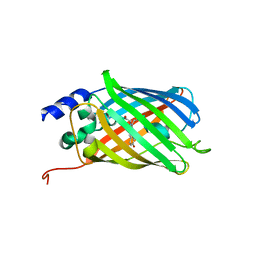 | | Crystal structure of rsEGFP2 in the fluorescent on-state | | Descriptor: | Green fluorescent protein | | Authors: | Adam, V, Martins, A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of ultrastable and reversibly photoswitchable fluorescent proteins for super-resolution imaging of the bacterial periplasm.
Sci Rep, 6, 2016
|
|
5DTY
 
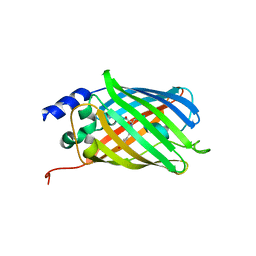 | |
7AV4
 
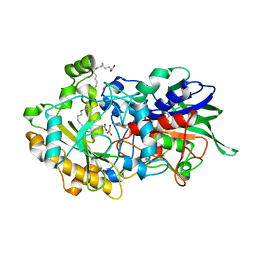 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6FG4
 
 | |
