1IHV
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | HIV-1 INTEGRASE | | 著者 | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | 登録日 | 1995-05-12 | | 公開日 | 1996-10-14 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
2EZC
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
1VKR
 
 | |
1VSQ
 
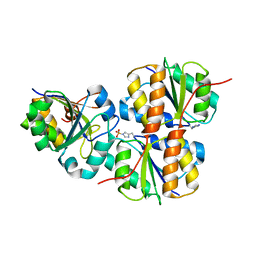 | |
2STW
 
 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | 著者 | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | 登録日 | 1996-08-05 | | 公開日 | 1997-03-12 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
2STT
 
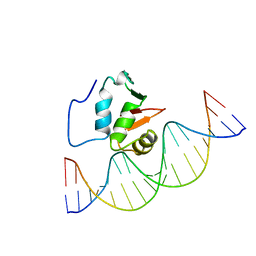 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, 25 STRUCTURES | | 分子名称: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | 著者 | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | 登録日 | 1996-08-05 | | 公開日 | 1997-03-12 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
2BBN
 
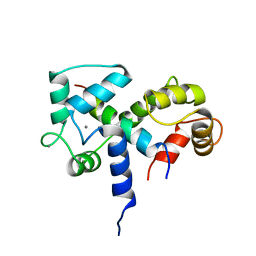 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2BBM
 
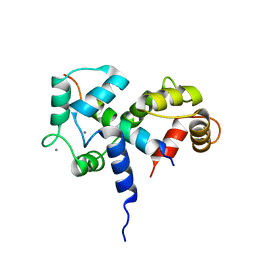 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2EZX
 
 | |
2EZY
 
 | |
2EZZ
 
 | |
2HIR
 
 | |
1WJB
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJA
 
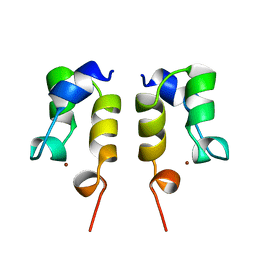 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
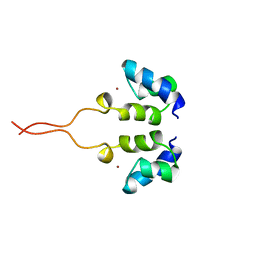 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJC
 
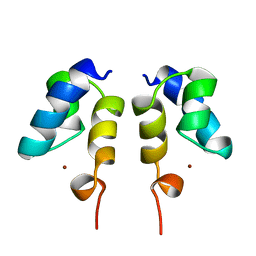 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1YUJ
 
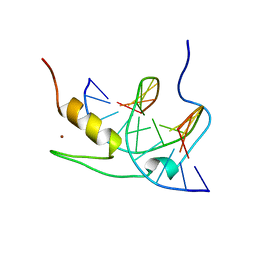 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | 著者 | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | 登録日 | 1996-12-31 | | 公開日 | 1997-12-31 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUI
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | 分子名称: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | 著者 | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | 登録日 | 1996-12-31 | | 公開日 | 1997-12-31 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
2CBH
 
 | |
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | TRANSPOSASE | | 著者 | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | 登録日 | 1997-07-25 | | 公開日 | 1997-12-03 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZF
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
