5XHP
 
 | | Transferase with ligands | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J, Yoo, Y, Kim, Y.H, Cho, H.S. | | Deposit date: | 2017-04-22 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of L-arginine and UDP bounded glycosyltransfease
To Be Published
|
|
5XYK
 
 | | Structure of Transferase | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J, Cho, H.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Transferase
To Be Published
|
|
2C83
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2C84
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
1J56
 
 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
5FJG
 
 | | The crystal structure of light-driven chloride pump ClR in pH 4.5. | | Descriptor: | ANHYDRORETINOL, CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Light-Driven Chloride Pump Clr in Ph 4.5.
To be Published
|
|
6J98
 
 | |
7YCO
 
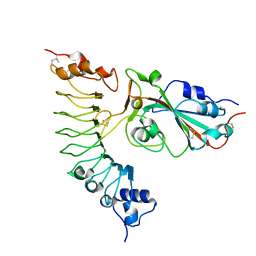 | |
7Y6K
 
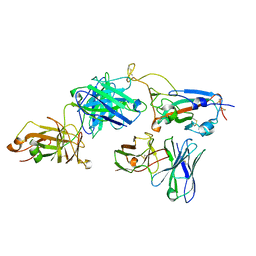 | |
7YM5
 
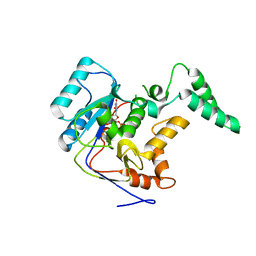 | |
7YM7
 
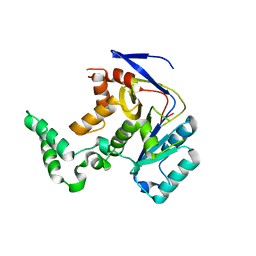 | |
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
7D85
 
 | |
2IY7
 
 | | crystal structure of the sialyltransferase PM0188 with CMP-3FNeuAc | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, LPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2006-07-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2IY8
 
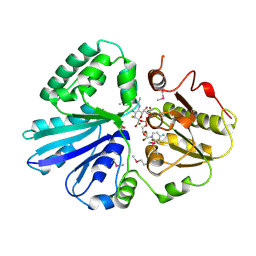 | | Crystal structure of the sialyltransferase PM0188 with CMP-3FNeuAc and lactose | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, PROTEIN PM0188, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2JGU
 
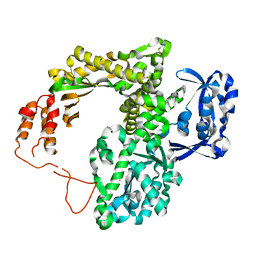 | | crystal structure of DNA-directed DNA polymerase | | Descriptor: | DNA POLYMERASE, MANGANESE (II) ION | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2007-02-15 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pfu, the High Fidelity DNA Polymerase from Pyrococcus Furiosus.
Int.J.Biol.Macromol., 42, 2008
|
|
1I9F
 
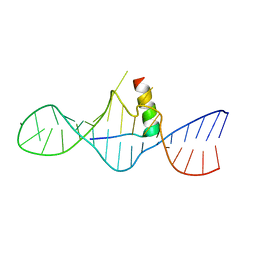 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
1L7M
 
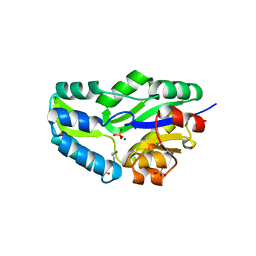 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7P
 
 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
2PLN
 
 | |
1FO5
 
 | | SOLUTION STRUCTURE OF REDUCED MJ0307 | | Descriptor: | THIOREDOXIN | | Authors: | Cave, J.W, Cho, H.S, Batchelder, A.M, Kim, R, Yokota, H, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-08-24 | | Release date: | 2001-04-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii.
Protein Sci., 10, 2001
|
|
