3WFN
 
 | |
7E0W
 
 | | Crystal Structure of BCH domain from S. pombe | | Descriptor: | Putative Rho GTPase-activating protein C1565.02c, TETRAETHYLENE GLYCOL | | Authors: | Chichili, V.P.R, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel intertwined anti-parallel dimeric structure of scaffold BCH domain regulates RhoA and RhoGAP functions
Proc.Natl.Acad.Sci.USA, 2021
|
|
4HEX
 
 | |
5BN5
 
 | | Structural basis for a unique ATP synthase core complex from Nanoarcheaum equitans | | Descriptor: | NEQ263, SULFATE ION, V-type ATP synthase alpha chain | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN3
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BN4
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ANP from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5BO5
 
 | | Structure of a unique ATP synthase subunit NeqB from Nanoarcheaum equitans | | Descriptor: | MAGNESIUM ION, NEQ263, SULFATE ION | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
8K70
 
 | |
4U00
 
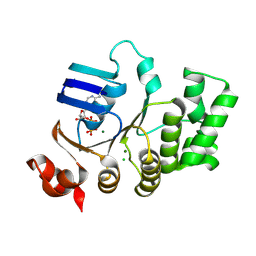 | | Crystal structure of TTHA1159 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Amino acid ABC transporter, ATP-binding protein, ... | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
4U02
 
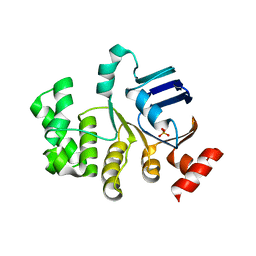 | | Crystal structure of apo-TTHA1159 | | Descriptor: | Amino acid ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
4E53
 
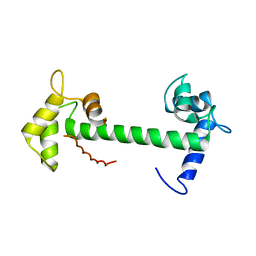 | | Calmodulin and Nm peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neuromodulin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
4E50
 
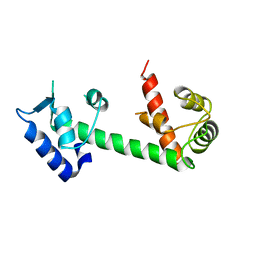 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
