5LT0
 
 | |
5LT2
 
 | |
5LT4
 
 | |
5LT3
 
 | |
5LT1
 
 | |
4LNU
 
 | | Nucleotide-free kinesin motor domain in complex with tubulin and a DARPin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Designed ankyrin repeat protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Cao, L, Gigant, B, Knossow, M. | | Deposit date: | 2013-07-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement
Nat Commun, 5, 2014
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
8KI9
 
 | |
8KIA
 
 | | Tomato spotted wilt virus L protein (apo state) | | Descriptor: | RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI8
 
 | | structure of Tomato spotted wilt virus L protein binding to 5'vRNA | | Descriptor: | RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI7
 
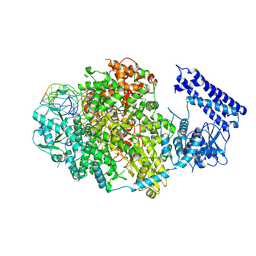 | | Structure of Tomato spotted wilt virus L protein contained endoH domain binding to 3'5'vRNA | | Descriptor: | RNA (5'-R(P*AP*CP*CP*UP*GP*AP*UP*UP*GP*CP*UP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3'), RNA (5'-R(P*GP*CP*AP*AP*UP*CP*AP*GP*G)-3'), ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
8KI6
 
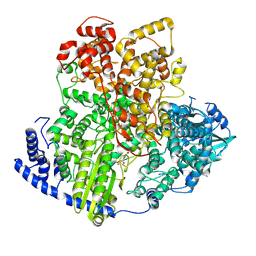 | | Structure of tomato spotted wilt virus L protein binding to Ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
7N3T
 
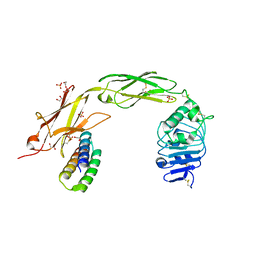 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
7S5B
 
 | |
7LCW
 
 | | Aspartimidylated Lasso Peptide Lihuanodin | | Descriptor: | Lasso Peptide Lihuanodin | | Authors: | Link, A.J, Cao, L. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cellulonodin-2 and Lihuanodin: Lasso Peptides with an Aspartimide Post-Translational Modification.
J.Am.Chem.Soc., 143, 2021
|
|
9J8V
 
 | | TSWV L protein in complex with ribavirin 5-triphosphate | | Descriptor: | RIBAVIRIN TRIPHOSPHATE, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2024-08-21 | | Release date: | 2025-04-16 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
6ADL
 
 | |
6ADR
 
 | |
6ADS
 
 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADT
 
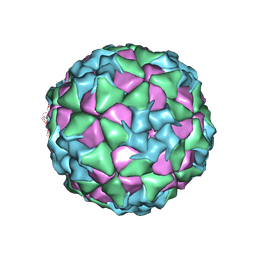 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADM
 
 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
