3CYZ
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with 9-keto-2(E)-decenoic acid at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CZ0
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CZ2
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera at pH 7.0 | | Descriptor: | CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
1O89
 
 | | Crystal structure of E. COLI K-12 yhdH | | Descriptor: | YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1O8C
 
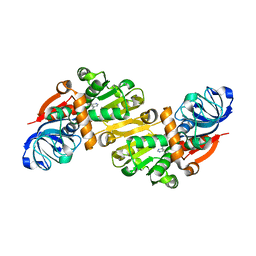 | | CRYSTAL STRUCTURE OF E. COLI K-12 YHDH WITH BOUND NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OJ7
 
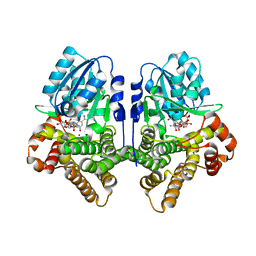 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
2FAV
 
 | |
2F0C
 
 | |
2FSD
 
 | |
2WKD
 
 | |
2WKC
 
 | |
